Dihydroceramide desaturase 1 (DES1) promotes anchorage-independent survival downstream of HER2-driven glucose uptake and metabolism
- PMID: 36165222
- PMCID: PMC9597949
- DOI: 10.1096/fj.202200748R
Dihydroceramide desaturase 1 (DES1) promotes anchorage-independent survival downstream of HER2-driven glucose uptake and metabolism
Abstract
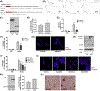
Oncogenic reprogramming of cellular metabolism is a hallmark of many cancers, but our mechanistic understanding of how such dysregulation is linked to tumor behavior remains poor. In this study, we have identified dihydroceramide desaturase (DES1)-which catalyzes the last step in de novo sphingolipid synthesis-as necessary for the acquisition of anchorage-independent survival (AIS), a key cancer enabling biology, and establish DES1 as a downstream effector of HER2-driven glucose uptake and metabolism. We further show that DES1 is sufficient to drive AIS and in vitro tumorigenicity and that increased DES1 levels-found in a third of HER2+ breast cancers-are associated with worse survival outcomes. Taken together, our findings reveal a novel pro-tumor role for DES1 as a transducer of HER2-driven glucose metabolic signals and provide evidence that targeting DES1 is an effective approach for overcoming AIS. Results further suggest that DES1 may have utility as a biomarker of aggressive and metastasis-prone HER2+ breast cancer.
Keywords: breast cancer; cell survival; metastasis; oncogene; sphingolipid.
© 2022 Federation of American Societies for Experimental Biology.
Conflict of interest statement
DISCLOSURES
The authors have stated explicitly that there are no conflicts of interest in connection with this article.
Figures
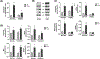
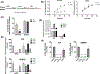




Similar articles
-
Myristic acid increases the activity of dihydroceramide Delta4-desaturase 1 through its N-terminal myristoylation.Biochimie. 2007 Dec;89(12):1553-61. doi: 10.1016/j.biochi.2007.07.001. Epub 2007 Jul 15. Biochimie. 2007. PMID: 17716801
-
Ablation of dihydroceramide desaturase confers resistance to etoposide-induced apoptosis in vitro.PLoS One. 2012;7(9):e44042. doi: 10.1371/journal.pone.0044042. Epub 2012 Sep 11. PLoS One. 2012. PMID: 22984457 Free PMC article.
-
Sphingolipid imbalance and inflammatory effects induced by uremic toxins in heart and kidney cells are reversed by dihydroceramide desaturase 1 inhibition.Toxicol Lett. 2021 Oct 10;350:133-142. doi: 10.1016/j.toxlet.2021.07.012. Epub 2021 Jul 22. Toxicol Lett. 2021. PMID: 34303789
-
Inhibitors of dihydroceramide desaturase 1: Therapeutic agents and pharmacological tools to decipher the role of dihydroceramides in cell biology.Chem Phys Lipids. 2016 May;197:33-44. doi: 10.1016/j.chemphyslip.2015.07.025. Epub 2015 Aug 3. Chem Phys Lipids. 2016. PMID: 26248324 Review.
-
Dihydroceramide desaturase and dihydrosphingolipids: debutant players in the sphingolipid arena.Prog Lipid Res. 2012 Apr;51(2):82-94. doi: 10.1016/j.plipres.2011.12.002. Epub 2011 Dec 17. Prog Lipid Res. 2012. PMID: 22200621 Review.
Cited by
-
Association Between 18F-FDG PET Activity and HER2 Status in Breast Cancer Brain Metastases.Nucl Med Mol Imaging. 2024 May;58(3):113-119. doi: 10.1007/s13139-024-00843-8. Epub 2024 Feb 1. Nucl Med Mol Imaging. 2024. PMID: 38633284 Free PMC article.
-
The Critical Impact of Sphingolipid Metabolism in Breast Cancer Progression and Drug Response.Int J Mol Sci. 2023 Jan 20;24(3):2107. doi: 10.3390/ijms24032107. Int J Mol Sci. 2023. PMID: 36768427 Free PMC article. Review.
References
-
- Debnath J, Mills KR, Collins NL, Reginato MJ, Muthuswamy SK, Brugge JS. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111(1):29–40. - PubMed
-
- Hawk MA, Gorsuch CL, Fagan P, et al. RIPK1-mediated induction of mitophagy compromises the viability of extracellular-matrix-detached cells. Nat Cell Biol. 2018;20(3): 272–284. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

