Kinetics of phagosome maturation is coupled to their intracellular motility
- PMID: 36163370
- PMCID: PMC9512794
- DOI: 10.1038/s42003-022-03988-4
Kinetics of phagosome maturation is coupled to their intracellular motility
Abstract
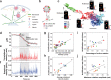
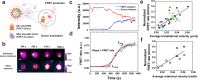
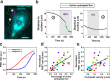
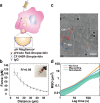
Immune cells degrade internalized pathogens in phagosomes through sequential biochemical changes. The degradation must be fast enough for effective infection control. The presumption is that each phagosome degrades cargos autonomously with a distinct but stochastic kinetic rate. However, here we show that the degradation kinetics of individual phagosomes is not stochastic but coupled to their intracellular motility. By engineering RotSensors that are optically anisotropic, magnetic responsive, and fluorogenic in response to degradation activities in phagosomes, we monitored cargo degradation kinetics in single phagosomes simultaneously with their translational and rotational dynamics. We show that phagosomes that move faster centripetally are more likely to encounter and fuse with lysosomes, thereby acidifying faster and degrading cargos more efficiently. The degradation rates increase nearly linearly with the translational and rotational velocities of phagosomes. Our results indicate that the centripetal motion of phagosomes functions as a clock for controlling the progression of cargo degradation.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Single-phagosome imaging reveals that homotypic fusion impairs phagosome degradative function.Biophys J. 2022 Feb 1;121(3):459-469. doi: 10.1016/j.bpj.2021.12.032. Epub 2021 Dec 29. Biophys J. 2022. PMID: 34968424 Free PMC article.
-
Quantitative Immunofluorescence to Study Phagosome Maturation and Resolution.Methods Mol Biol. 2023;2692:121-137. doi: 10.1007/978-1-0716-3338-0_9. Methods Mol Biol. 2023. PMID: 37365465
-
Phagosome resolution regenerates lysosomes and maintains the degradative capacity in phagocytes.J Cell Biol. 2021 Sep 6;220(9):e202005072. doi: 10.1083/jcb.202005072. Epub 2021 Jun 28. J Cell Biol. 2021. PMID: 34180943 Free PMC article.
-
A guide to measuring phagosomal dynamics.FEBS J. 2021 Mar;288(5):1412-1433. doi: 10.1111/febs.15506. Epub 2020 Aug 16. FEBS J. 2021. PMID: 32757358 Free PMC article. Review.
-
Phagosome maturation and modulation of macrophage effector function by intracellular pathogens: target for therapeutics.Future Microbiol. 2022 Jan;17:59-76. doi: 10.2217/fmb-2021-0101. Epub 2021 Dec 8. Future Microbiol. 2022. PMID: 34877879 Review.
Cited by
-
Propulsive cell entry diverts pathogens from immune degradation by remodeling the phagocytic synapse.Proc Natl Acad Sci U S A. 2023 Dec 5;120(49):e2306788120. doi: 10.1073/pnas.2306788120. Epub 2023 Nov 30. Proc Natl Acad Sci U S A. 2023. PMID: 38032935 Free PMC article.
-
Nanoparticles for Interrogation of Cell Signaling.Annu Rev Anal Chem (Palo Alto Calif). 2023 Jun 14;16(1):333-351. doi: 10.1146/annurev-anchem-092822-085852. Annu Rev Anal Chem (Palo Alto Calif). 2023. PMID: 37314874 Free PMC article. Review.
-
Propulsive cell entry diverts pathogens from immune degradation by remodeling the phagocytic synapse.bioRxiv [Preprint]. 2023 Apr 28:2023.04.25.538287. doi: 10.1101/2023.04.25.538287. bioRxiv. 2023. Update in: Proc Natl Acad Sci U S A. 2023 Dec 5;120(49):e2306788120. doi: 10.1073/pnas.2306788120 PMID: 37162866 Free PMC article. Updated. Preprint.
-
Identification of the effects of alkalinity exposure on the gills of oriental river prawns, Macrobrachium nipponense.BMC Genomics. 2024 Aug 6;25(1):765. doi: 10.1186/s12864-024-10659-7. BMC Genomics. 2024. PMID: 39107708 Free PMC article.
-
epHero - a tandem-fluorescent probe to track the fate of apoptotic cells during efferocytosis.Cell Death Discov. 2024 Apr 17;10(1):179. doi: 10.1038/s41420-024-01952-1. Cell Death Discov. 2024. PMID: 38632247 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

