Rapid diagnosis of Talaromyces marneffei infection by metagenomic next-generation sequencing technology in a Chinese cohort of inborn errors of immunity
- PMID: 36159645
- PMCID: PMC9493038
- DOI: 10.3389/fcimb.2022.987692
Rapid diagnosis of Talaromyces marneffei infection by metagenomic next-generation sequencing technology in a Chinese cohort of inborn errors of immunity
Abstract
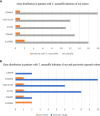
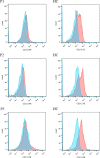
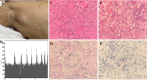
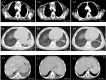
Talaromyces marneffei (T. marneffei) is an opportunistic pathogen. Patients with inborn errors of immunity (IEI) have been increasingly diagnosed with T. marneffei in recent years. The disseminated infection of T. marneffei can be life-threatening without timely and effective antifungal therapy. Rapid and accurate pathogenic microbiological diagnosis is particularly critical for these patients. A total of 505 patients with IEI were admitted to our hospital between January 2019 and June 2022, among whom T. marneffei was detected in 6 patients by metagenomic next-generation sequencing (mNGS), and their clinical and immunological characteristics were summarized. We performed a systematic literature review on T. marneffei infections with published immunodeficiency-related gene mutations. All patients in our cohort were confirmed to have genetic mutations in IL12RB1, IFNGR1, STAT1, STAT3, and CD40LG. T. marneffei was detected in both the blood and lymph nodes of P1 with IL12RB1 mutations, and the clinical manifestations were serious and included recurrent fever, weight loss, severe anemia, splenomegaly and lymphadenopathy, all requiring long-term antifungal therapy. These six patients received antifungal treatment, which relieved symptoms and improved imaging findings. Five patients survived, while one patient died of sepsis after hematopoietic stem cell transplantation. The application of mNGS methods for pathogen detection in IEI patients and comparison with traditional diagnosis methods were investigated. Traditional diagnostic methods and mNGS tests were performed simultaneously in 232 patients with IEI. Compared to the traditional methods, the sensitivity and specificity of mNGS in diagnosing T. marneffei infection were 100% and 98.7%, respectively. The reporting time for T. marneffei detection was approximately 26 hours by mNGS, 3-14 days by culture, and 6-11 days by histopathology. T. marneffei infection was first reported in IEI patients with IL12RB1 gene mutation, which expanded the IEI lineage susceptible to T. marneffei. For IEI patients with T. marneffei infection, we highlight the application of mNGS in pathogenic detection. mNGS is recommended as a front-line diagnostic test for rapidly identifying pathogens in complex and severe infections.
Keywords: IL12RB1 mutation; T-cell-mediated immunity; Talaromyces marneffei; inborn errors of immunity; intrinsic and innate immunodeficiencies; metagenomic next-generation sequencing.
Copyright © 2022 Liu, Sun, Ying, Liu, Wang, Sun, Wang, Yang, Hui, Zhou, Hou and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Case report: Diagnosis of Talaromyces marneffei infection in an HIV-negative patient with septic shock and high-titer anti-interferon gamma autoantibodies by metagenomic next-generation sequencing.Front Cell Infect Microbiol. 2023 Jul 4;13:1163846. doi: 10.3389/fcimb.2023.1163846. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37469600 Free PMC article.
-
Clinical usefulness of metagenomic next-generation sequencing for Talaromyces marneffei diagnosis in China: a retrospective study.Eur J Clin Microbiol Infect Dis. 2024 Jul;43(7):1367-1374. doi: 10.1007/s10096-024-04856-1. Epub 2024 May 27. Eur J Clin Microbiol Infect Dis. 2024. PMID: 38801485
-
Unusual disseminated Talaromyces marneffei infection mimicking lymphoma in a non-immunosuppressed patient in East China: a case report and review of the literature.BMC Infect Dis. 2020 Oct 28;20(1):800. doi: 10.1186/s12879-020-05526-1. BMC Infect Dis. 2020. PMID: 33115429 Free PMC article. Review.
-
Clinical features of patients with talaromycosis marneffei and microbiological characteristics of the causative strains.J Clin Lab Anal. 2022 Nov;36(11):e24737. doi: 10.1002/jcla.24737. Epub 2022 Oct 21. J Clin Lab Anal. 2022. PMID: 36268985 Free PMC article.
-
Cerebral vasculitis caused by Talaromyces marneffei and Aspergillus niger in a HIV-positive patient: a case report and literature review.J Neurovirol. 2022 Apr;28(2):274-280. doi: 10.1007/s13365-021-01032-5. Epub 2022 Jan 3. J Neurovirol. 2022. PMID: 34981436 Free PMC article. Review.
Cited by
-
Utilizing metagenomic next-generation sequencing for pathogen detection and diagnosis in lower respiratory tract infections in real-world clinical practice.Infection. 2024 Apr;52(2):625-636. doi: 10.1007/s15010-024-02185-1. Epub 2024 Feb 18. Infection. 2024. PMID: 38368306
-
Disseminated Combined Talaromyces marneffei and Enterococcus faecium Bloodstream Infection Presenting as Gastrointestinal Perforation in a Patient with CARD9 Gene Mutation.Infect Drug Resist. 2024 Oct 31;17:4783-4790. doi: 10.2147/IDR.S479629. eCollection 2024. Infect Drug Resist. 2024. PMID: 39498413 Free PMC article.
-
Case report: Diagnosis of Talaromyces marneffei infection in an HIV-negative patient with septic shock and high-titer anti-interferon gamma autoantibodies by metagenomic next-generation sequencing.Front Cell Infect Microbiol. 2023 Jul 4;13:1163846. doi: 10.3389/fcimb.2023.1163846. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37469600 Free PMC article.
-
Disseminated Talaromyces marneffei infection in a child with HIV infection.J Hematop. 2023 Sep;16(3):181-183. doi: 10.1007/s12308-023-00551-w. Epub 2023 Jun 15. J Hematop. 2023. PMID: 38175399 No abstract available.
-
Characteristics of Endemic Mycoses Talaromyces marneffei Infection Associated with Inborn Errors of Immunity.J Clin Immunol. 2024 Sep 26;45(1):17. doi: 10.1007/s10875-024-01798-3. J Clin Immunol. 2024. PMID: 39325235
References
-
- Cao Q., Chen B., Chen L., Chen P., Bijie H. (2020). Expert consensus on the clinical application of metagenomics next-generation sequencing technology for detection of infectious pathogens in China. Chin. J. Infect. Dis. 38 (11), 681–689. doi: 10.3760/cma.j.cn311365-20200731-00732 - DOI
-
- Cao C., Xi L., Chaturvedi V. (2019). Talaromycosis (Penicilliosis) due to talaromyces (Penicillium) marneffei: Insights into the clinical trends of a major fungal disease 60 years after the discovery of the pathogen. MYCOPATHOL [Journal Article; Review]. 184 (6), 709–720. doi: 10.1007/s11046-019-00410-2 - DOI - PubMed
-
- Chen Z., Cheng H., Cai Z., Wei Q., Li J., Liang J., et al. . (2021). Identification of microbiome etiology associated with drug resistance in pleural empyema. Front. Cell Infect. Microbiol. [Journal Article; Res. Support Non-U.S. Gov't]. 11, 637018. doi: 10.3389/fcimb.2021.637018 - DOI - PMC - PubMed
-
- Chen K., Tan J., Qian S., Wu S., Chen Q. (2021). Case report: Disseminated talaromyces marneffei infection in a patient with chronic mucocutaneous candidiasis and a novel STAT1 gain-of-Function mutation. Front. Immunol. [Case Reports; Res. Support Non-U.S. Gov't]. 12, 682350. doi: 10.3389/fimmu.2021.682350 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

