Chaperone-assisted E3 ligase CHIP: A double agent in cancer
- PMID: 36157498
- PMCID: PMC9485218
- DOI: 10.1016/j.gendis.2021.08.003
Chaperone-assisted E3 ligase CHIP: A double agent in cancer
Abstract
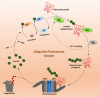
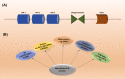
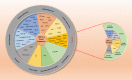
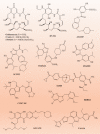
The carboxy-terminus of Hsp70-interacting protein (CHIP) is a ubiquitin ligase and co-chaperone belonging to Ubox family that plays a crucial role in the maintenance of cellular homeostasis by switching the equilibrium of the folding-refolding mechanism towards the proteasomal or lysosomal degradation pathway. It links molecular chaperones viz. HSC70, HSP70 and HSP90 with ubiquitin proteasome system (UPS), acting as a quality control system. CHIP contains charged domain in between N-terminal tetratricopeptide repeat (TPR) and C-terminal Ubox domain. TPR domain interacts with the aberrant client proteins via chaperones while Ubox domain facilitates the ubiquitin transfer to the client proteins for ubiquitination. Thus, CHIP is a classic molecule that executes ubiquitination for degradation of client proteins. Further, CHIP has been found to be indulged in cellular differentiation, proliferation, metastasis and tumorigenesis. Additionally, CHIP can play its dual role as a tumor suppressor as well as an oncogene in numerous malignancies, thus acting as a double agent. Here, in this review, we have reported almost all substrates of CHIP established till date and classified them according to the hallmarks of cancer. In addition, we discussed about its architectural alignment, tissue specific expression, sub-cellular localization, folding-refolding mechanisms of client proteins, E4 ligase activity, normal physiological roles, as well as involvement in various diseases and tumor biology. Further, we aim to discuss its importance in HSP90 inhibitors mediated cancer therapy. Thus, this report concludes that CHIP may be a promising and worthy drug target towards pharmaceutical industry for drug development.
Keywords: CHIP; Chaperones (HSC70/HSP70 & HSP90); Oncogene; Therapy; Tumor suppressor; Ubiquitin proteasome system (UPS).
© 2021 Chongqing Medical University. Production and hosting by Elsevier B.V.
Figures


Similar articles
-
CHIP: A Co-chaperone for Degradation by the Proteasome and Lysosome.Subcell Biochem. 2023;101:351-387. doi: 10.1007/978-3-031-14740-1_12. Subcell Biochem. 2023. PMID: 36520313
-
CHIP: a co-chaperone for degradation by the proteasome.Subcell Biochem. 2015;78:219-42. doi: 10.1007/978-3-319-11731-7_11. Subcell Biochem. 2015. PMID: 25487024 Review.
-
CHIP: a quality-control E3 ligase collaborating with molecular chaperones.Int J Biochem Cell Biol. 2003 May;35(5):572-8. doi: 10.1016/s1357-2725(02)00394-1. Int J Biochem Cell Biol. 2003. PMID: 12672450 Review.
-
Ca2+/S100 proteins act as upstream regulators of the chaperone-associated ubiquitin ligase CHIP (C terminus of Hsc70-interacting protein).J Biol Chem. 2013 Mar 8;288(10):7158-68. doi: 10.1074/jbc.M112.436758. Epub 2013 Jan 23. J Biol Chem. 2013. PMID: 23344957 Free PMC article.
-
The ubiquitin ligase CHIP regulates c-Myc stability and transcriptional activity.Oncogene. 2013 Mar 7;32(10):1284-95. doi: 10.1038/onc.2012.144. Epub 2012 Apr 30. Oncogene. 2013. PMID: 22543587
Cited by
-
Early 2-Factor Transcription Factors Associated with Progression and Recurrence in Bevacizumab-Responsive Subtypes of Glioblastoma.Cancers (Basel). 2024 Jul 14;16(14):2536. doi: 10.3390/cancers16142536. Cancers (Basel). 2024. PMID: 39061176 Free PMC article.
-
Control of SOX2 protein stability and tumorigenic activity by E3 ligase CHIP in esophageal cancer cells.Oncogene. 2023 Jul;42(30):2315-2328. doi: 10.1038/s41388-023-02745-z. Epub 2023 Jun 23. Oncogene. 2023. PMID: 37353616
-
COVID-19 therapeutics: Clinical application of repurposed drugs and futuristic strategies for target-based drug discovery.Genes Dis. 2023 Jul;10(4):1402-1428. doi: 10.1016/j.gendis.2022.12.019. Epub 2023 Apr 7. Genes Dis. 2023. PMID: 37334160 Free PMC article. Review.
-
Chaperone-mediated autophagy modulates Snail protein stability: implications for breast cancer metastasis.Mol Cancer. 2024 Oct 11;23(1):227. doi: 10.1186/s12943-024-02138-0. Mol Cancer. 2024. PMID: 39390584 Free PMC article.
-
Isoform alterations in the ubiquitination machinery impacting gastrointestinal malignancies.Cell Death Dis. 2024 Mar 8;15(3):194. doi: 10.1038/s41419-024-06575-z. Cell Death Dis. 2024. PMID: 38453895 Free PMC article. Review.
References
-
- Broadley S.A., Hartl F.U. The role of molecular chaperones in human misfolding diseases. FEBS Lett. 2009;583(16):2647–2653. - PubMed
-
- Brodsky J.L., Chiosis G. Hsp70 molecular chaperones: emerging roles in human disease and identification of small molecule modulators. Curr Top Med Chem. 2006;6(11):1215–1225. - PubMed
-
- Ellis R.J., Hemmingsen S.M. Molecular chaperones: proteins essential for the biogenesis of some macromolecular structures. Trends Biochem Sci. 1989;14(8):339–342. - PubMed
-
- Martin J., Hartl F.U. Molecular chaperones in cellular protein folding. Bioessays. 1994;16(9):689–692. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

