ATF1 Restricts Human Herpesvirus 6A Replication via Beta Interferon Induction
- PMID: 36154610
- PMCID: PMC9555150
- DOI: 10.1128/jvi.01264-22
ATF1 Restricts Human Herpesvirus 6A Replication via Beta Interferon Induction
Abstract
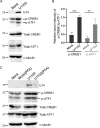
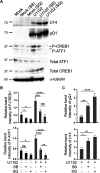
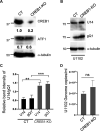
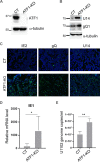
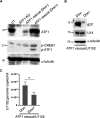
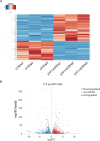
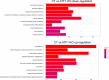
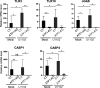
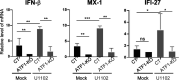
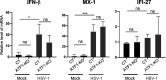
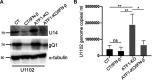
The stimulus-induced cAMP response element (CRE)-binding protein (CREB) family of transcription factors bind to CREs to regulate diverse cellular responses, including proliferation, survival, and differentiation. Human herpesvirus 6A (HHV-6A), which belongs to the Betaherpesvirinae subfamily, is a lymphotropic herpesvirus frequently found in patients with neuroinflammatory diseases. Previous reports implicated the importance of CREs in the HHV-6A life cycle, although the effects of the binding of transcription factors to CREs in viral replication have not been fully elucidated. In this study, we analyzed the role of the CREB family of transcription factors during HHV-6A replication. We found that HHV-6A infection enhanced phosphorylation of the CREB family members CREB1 and activating transcription factor 1 (ATF1). Knockout (KO) of CREB1 or ATF1 enhanced viral gene expression and viral replication. The increase in viral yields in supernatants from ATF1-KO cells was greater than that in supernatants from CREB1-KO cells. Transcriptome sequencing (RNA-seq) analysis showed that sensors of the innate immune system were downregulated in ATF1-KO cells, and mRNAs of beta interferon (IFN-β) and IFN-regulated genes were reduced in these cells infected with HHV-6A. IFN-β treatment of ATF1-KO cells reduced progeny viral yields significantly, suggesting that the enhancement of viral replication was caused by a reduction of IFN-β. Taken together, our results suggest that ATF1 is activated during HHV-6A infection and restricts viral replication via IFN-β induction. IMPORTANCE Human herpesvirus 6A (HHV-6A) is a ubiquitous herpesvirus implicated in Alzheimer's disease, although its role in its pathogenesis has not been confirmed. Here, we showed that the transcription factor ATF1 restricts HHV-6A replication, mediated by IFN-β induction. Our study provides new insights into the role of ATF1 in innate viral immunity and reveals the importance of IFN-β for regulation of HHV-6A replication, which possibly impairs HHV-6A pathogenesis.
Keywords: ATF1; HHV-6; gene expression; herpesviruses; interferons; transcription factors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Human herpesvirus 6A nuclear matrix protein U37 interacts with heat shock transcription factor 1 and activates the heat shock response.J Virol. 2023 Sep 28;97(9):e0071823. doi: 10.1128/jvi.00718-23. Epub 2023 Sep 6. J Virol. 2023. PMID: 37671864 Free PMC article.
-
Human Herpesvirus 6A Tegument Protein U14 Induces NF-κB Signaling by Interacting with p65.J Virol. 2021 Nov 9;95(23):e0126921. doi: 10.1128/JVI.01269-21. Epub 2021 Sep 22. J Virol. 2021. PMID: 34549982 Free PMC article.
-
A human herpesvirus 6A-encoded microRNA: role in viral lytic replication.J Virol. 2015 Mar;89(5):2615-27. doi: 10.1128/JVI.02007-14. Epub 2014 Dec 17. J Virol. 2015. PMID: 25520507 Free PMC article.
-
Molecular biology of human herpesviruses 6A and 6B.Infect Agents Dis. 1993 Dec;2(6):343-60. Infect Agents Dis. 1993. PMID: 8012736 Review.
-
[Human herpesvirus 6 A enhances HIV-1 replication in vitro by soluble mediators, this effect is diminished by endotoxin].Orv Hetil. 1999 Nov 14;140(46):2577-80. Orv Hetil. 1999. PMID: 10628199 Review. Hungarian.
Cited by
-
Human herpesvirus 6A nuclear matrix protein U37 interacts with heat shock transcription factor 1 and activates the heat shock response.J Virol. 2023 Sep 28;97(9):e0071823. doi: 10.1128/jvi.00718-23. Epub 2023 Sep 6. J Virol. 2023. PMID: 37671864 Free PMC article.
References
-
- Aubin JT, Collandre H, Candotti D, Ingrand D, Rouzioux C, Burgard M, Richard S, Huraux JM, Agut H. 1991. Several groups among human herpesvirus-6 strains can be distinguished by Southern blotting and polymerase chain-reaction. J Clin Microbiol 29:367–372. 10.1128/jcm.29.2.367-372.1991. - DOI - PMC - PubMed
-
- Ablashi D, Agut H, Alvarez-Lafuente R, Clark DA, Dewhurst S, DiLuca D, Flamand L, Frenkel N, Gallo R, Gompels UA, Hollsberg P, Jacobson S, Luppi M, Lusso P, Malnati M, Medveczky P, Mori Y, Pellett PE, Pritchett JC, Yamanishi K, Yoshikawa T. 2014. Classification of HHV-6A and HHV-6B as distinct viruses. Arch Virol 159:863–870. 10.1007/s00705-013-1902-5. - DOI - PMC - PubMed
-
- Yamanishi K, Mori Y, Pellet PE. 2013. Human herpesviruses 6 and 7, p 2058–2079. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed. Lippincott-Williams &Wilkins, Philadelphia, PA.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

