MERS-CoV nsp1 regulates autophagic flux via mTOR signalling and dysfunctional lysosomes
- PMID: 36153658
- PMCID: PMC9621213
- DOI: 10.1080/22221751.2022.2128434
MERS-CoV nsp1 regulates autophagic flux via mTOR signalling and dysfunctional lysosomes
Abstract
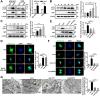
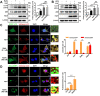
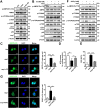
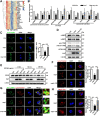
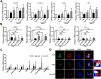
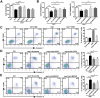
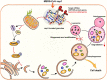
Autophagy, a cellular surveillance mechanism, plays an important role in combating invading pathogens. However, viruses have evolved various strategies to disrupt autophagy and even hijack it for replication and release. Here, we demonstrated that Middle East respiratory syndrome coronavirus (MERS-CoV) non-structural protein 1(nsp1) induces autophagy but inhibits autophagic activity. MERS-CoV nsp1 expression increased ROS and reduced ATP levels in cells, which activated AMPK and inhibited the mTOR signalling pathway, resulting in autophagy induction. Meanwhile, as an endonuclease, MERS-CoV nsp1 downregulated the mRNA of lysosome-related genes that were enriched in nsp1-located granules, which diminished lysosomal biogenesis and acidification, and inhibited autophagic flux. Importantly, MERS-CoV nsp1-induced autophagy can lead to cell death in vitro and in vivo. These findings clarify the mechanism by which MERS-CoV nsp1-mediated autophagy regulation, providing new insights for the prevention and treatment of the coronavirus.
Keywords: MERS-CoV; autophagic flux; lysosomes; mTOR; nsp1.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

Similar articles
-
The Endonucleolytic RNA Cleavage Function of nsp1 of Middle East Respiratory Syndrome Coronavirus Promotes the Production of Infectious Virus Particles in Specific Human Cell Lines.J Virol. 2018 Oct 12;92(21):e01157-18. doi: 10.1128/JVI.01157-18. Print 2018 Nov 1. J Virol. 2018. PMID: 30111568 Free PMC article.
-
MERS-CoV nsp1 impairs the cellular metabolic processes by selectively downregulating mRNAs in a novel granules.Virulence. 2022 Dec;13(1):355-369. doi: 10.1080/21505594.2022.2032928. Virulence. 2022. PMID: 35129074 Free PMC article.
-
Middle East Respiratory Syndrome Coronavirus nsp1 Inhibits Host Gene Expression by Selectively Targeting mRNAs Transcribed in the Nucleus while Sparing mRNAs of Cytoplasmic Origin.J Virol. 2015 Nov;89(21):10970-81. doi: 10.1128/JVI.01352-15. Epub 2015 Aug 26. J Virol. 2015. PMID: 26311885 Free PMC article.
-
The lysosome: a crucial hub for AMPK and mTORC1 signalling.Biochem J. 2017 Apr 13;474(9):1453-1466. doi: 10.1042/BCJ20160780. Biochem J. 2017. PMID: 28408430 Review.
-
Mechanisms of Coronavirus Nsp1-Mediated Control of Host and Viral Gene Expression.Cells. 2021 Feb 2;10(2):300. doi: 10.3390/cells10020300. Cells. 2021. PMID: 33540583 Free PMC article. Review.
Cited by
-
The relationship between autophagy and respiratory viruses.Arch Microbiol. 2024 Mar 4;206(4):136. doi: 10.1007/s00203-024-03838-3. Arch Microbiol. 2024. PMID: 38436746 Review.
-
The role of reactive oxygen species in severe acute respiratory syndrome coronavirus 2 (SARS-COV-2) infection-induced cell death.Cell Mol Biol Lett. 2024 Nov 8;29(1):138. doi: 10.1186/s11658-024-00659-6. Cell Mol Biol Lett. 2024. PMID: 39516736 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
