Ablation of the endoplasmic reticulum stress kinase PERK induces paraptosis and type I interferon to promote anti-tumor T cell responses
- PMID: 36150390
- PMCID: PMC9561067
- DOI: 10.1016/j.ccell.2022.08.016
Ablation of the endoplasmic reticulum stress kinase PERK induces paraptosis and type I interferon to promote anti-tumor T cell responses
Abstract
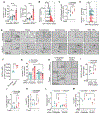
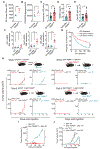
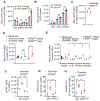
Activation of unfolded protein responses (UPRs) in cancer cells undergoing endoplasmic reticulum (ER) stress promotes survival. However, how UPR in tumor cells impacts anti-tumor immune responses remains poorly described. Here, we investigate the role of the UPR mediator pancreatic ER kinase (PKR)-like ER kinase (PERK) in cancer cells in the modulation of anti-tumor immunity. Deletion of PERK in cancer cells or pharmacological inhibition of PERK in melanoma-bearing mice incites robust activation of anti-tumor T cell immunity and attenuates tumor growth. PERK elimination in ER-stressed malignant cells triggers SEC61β-induced paraptosis, thereby promoting immunogenic cell death (ICD) and systemic anti-tumor responses. ICD induction in PERK-ablated tumors stimulates type I interferon production in dendritic cells (DCs), which primes CCR2-dependent tumor trafficking of common-monocytic precursors and their intra-tumor commitment into monocytic-lineage inflammatory Ly6C+CD103+ DCs. These findings identify how tumor cell-derived PERK promotes immune evasion and highlight the potential of PERK-targeting therapies in cancer immunotherapy.
Keywords: PERK; immunogenic cell death; tumor immunity; type I IFN; unfolded protein responses.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Similar articles
-
The PERK Arm of the Unfolded Protein Response Negatively Regulates Transmissible Gastroenteritis Virus Replication by Suppressing Protein Translation and Promoting Type I Interferon Production.J Virol. 2018 Jul 17;92(15):e00431-18. doi: 10.1128/JVI.00431-18. Print 2018 Aug 1. J Virol. 2018. PMID: 29769338 Free PMC article.
-
Attenuation of PKR-like ER Kinase (PERK) Signaling Selectively Controls Endoplasmic Reticulum Stress-induced Inflammation Without Compromising Immunological Responses.J Biol Chem. 2016 Jul 22;291(30):15830-40. doi: 10.1074/jbc.M116.738021. Epub 2016 May 23. J Biol Chem. 2016. PMID: 27226638 Free PMC article.
-
The Unfolded Protein Response Mediator PERK Governs Myeloid Cell-Driven Immunosuppression in Tumors through Inhibition of STING Signaling.Immunity. 2020 Apr 14;52(4):668-682.e7. doi: 10.1016/j.immuni.2020.03.004. Immunity. 2020. PMID: 32294407 Free PMC article.
-
Protein-rich foods, sea foods, and gut microbiota amplify immune responses in chronic diseases and cancers - Targeting PERK as a novel therapeutic strategy for chronic inflammatory diseases, neurodegenerative disorders, and cancer.Pharmacol Ther. 2024 Mar;255:108604. doi: 10.1016/j.pharmthera.2024.108604. Epub 2024 Feb 13. Pharmacol Ther. 2024. PMID: 38360205 Review.
-
Unfolding anti-tumor immunity: ER stress responses sculpt tolerogenic myeloid cells in cancer.J Immunother Cancer. 2017 Jan 17;5:5. doi: 10.1186/s40425-016-0203-4. eCollection 2017. J Immunother Cancer. 2017. PMID: 28105371 Free PMC article. Review.
Cited by
-
HKDC1 promotes tumor immune evasion in hepatocellular carcinoma by coupling cytoskeleton to STAT1 activation and PD-L1 expression.Nat Commun. 2024 Feb 13;15(1):1314. doi: 10.1038/s41467-024-45712-2. Nat Commun. 2024. PMID: 38351096 Free PMC article.
-
Exploring paraptosis as a therapeutic approach in cancer treatment.J Biomed Sci. 2024 Nov 4;31(1):101. doi: 10.1186/s12929-024-01089-4. J Biomed Sci. 2024. PMID: 39497143 Free PMC article. Review.
-
HRS mediates tumor immune evasion by regulating proteostasis-associated interferon pathway activation.Cell Rep. 2023 Nov 28;42(11):113352. doi: 10.1016/j.celrep.2023.113352. Epub 2023 Nov 9. Cell Rep. 2023. PMID: 37948180 Free PMC article.
-
ATF6 Promotes Colorectal Cancer Growth and Stemness by Regulating the Wnt Pathway.Cancer Res Commun. 2024 Oct 1;4(10):2734-2755. doi: 10.1158/2767-9764.CRC-24-0268. Cancer Res Commun. 2024. PMID: 39324706 Free PMC article.
-
Emerging dimensions of autophagy in melanoma.Autophagy. 2024 Aug;20(8):1700-1711. doi: 10.1080/15548627.2024.2330261. Epub 2024 Mar 21. Autophagy. 2024. PMID: 38497492 Review.
References
-
- Atkins C, Liu Q, Minthorn E, Zhang SY, Figueroa DJ, Moss K, Stanley TB, Sanders B , Goetz A, Gaul N, et al. (2013). Characterization of a novel PERK kinase inhibitor with antitumor and antiangiogenic activity. Cancer Res. 73, 1993–2002. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

