The Rise and Fall of SARS-CoV-2 Variants and Ongoing Diversification of Omicron
- PMID: 36146815
- PMCID: PMC9505243
- DOI: 10.3390/v14092009
The Rise and Fall of SARS-CoV-2 Variants and Ongoing Diversification of Omicron
Abstract
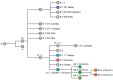
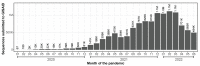
In late December of 2019, high-throughput sequencing technologies enabled rapid identification of SARS-CoV-2 as the etiological agent of COVID-19, and global sequencing efforts are now a critical tool for monitoring the ongoing spread and evolution of this virus. Here, we provide a short retrospective analysis of SARS-CoV-2 variants by analyzing a subset (n = 97,437) of all publicly available SARS-CoV-2 genomes (n = ~11.9 million) that were randomly selected but equally distributed over the course of the pandemic. We plot the appearance of new variants of concern (VOCs) over time and show that the mutation rates in Omicron (BA.1) and Omicron sub-lineages (BA.2-BA.5) are significantly elevated compared to previously identified SARS-CoV-2 variants. Mutations in Omicron are primarily restricted to the spike and nucleocapsid proteins, while 24 other viral proteins-including those involved in SARS-CoV-2 replication-are generally conserved. Collectively, this suggests that the genetic distinction of Omicron primarily arose from selective pressures on the spike, and that the fidelity of replication of this variant has not been altered.
Keywords: BA.4; BA.5; COVID-19; Omicron; SARS-CoV-2; viral surveillance.
Conflict of interest statement
B.W. is the founder of SurGene LLC, and VIRIS Detection Systems Inc. B.W., A. Nemudryi and A. Nemudraia, and A.S.-F. are inventors on patents related to CRISPR-Cas systems and applications thereof.
Figures

Similar articles
-
Accessible and Adaptable Multiplexed Real-Time PCR Approaches to Identify SARS-CoV-2 Variants of Concern.Microbiol Spectr. 2022 Oct 26;10(5):e0322222. doi: 10.1128/spectrum.03222-22. Epub 2022 Sep 15. Microbiol Spectr. 2022. PMID: 36106882 Free PMC article.
-
Tracking SARS-CoV-2 Omicron diverse spike gene mutations identifies multiple inter-variant recombination events.Signal Transduct Target Ther. 2022 Apr 26;7(1):138. doi: 10.1038/s41392-022-00992-2. Signal Transduct Target Ther. 2022. PMID: 35474215 Free PMC article.
-
Sustained applicability of SARS-CoV-2 variants identification by Sanger Sequencing Strategy on emerging various SARS-CoV-2 Omicron variants in Hiroshima, Japan.BMC Genomics. 2024 Nov 11;25(1):1063. doi: 10.1186/s12864-024-10973-0. BMC Genomics. 2024. PMID: 39528931 Free PMC article.
-
The outbreak of SARS-CoV-2 Omicron lineages, immune escape, and vaccine effectivity.J Med Virol. 2023 Jan;95(1):e28138. doi: 10.1002/jmv.28138. Epub 2022 Sep 21. J Med Virol. 2023. PMID: 36097349 Free PMC article. Review.
-
Omicron: What Makes the Latest SARS-CoV-2 Variant of Concern So Concerning?J Virol. 2022 Mar 23;96(6):e0207721. doi: 10.1128/jvi.02077-21. Epub 2022 Mar 23. J Virol. 2022. PMID: 35225672 Free PMC article. Review.
Cited by
-
Identifying Structural Features of Nucleotide Analogues to Overcome SARS-CoV-2 Exonuclease Activity.Viruses. 2022 Jun 28;14(7):1413. doi: 10.3390/v14071413. Viruses. 2022. PMID: 35891393 Free PMC article.
-
Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.iScience. 2024 Jan 19;27(2):108976. doi: 10.1016/j.isci.2024.108976. eCollection 2024 Feb 16. iScience. 2024. PMID: 38327783 Free PMC article.
-
Antigenicity comparison of SARS-CoV-2 Omicron sublineages with other variants contained multiple mutations in RBD.MedComm (2020). 2022 Apr 9;3(2):e130. doi: 10.1002/mco2.130. eCollection 2022 Jun. MedComm (2020). 2022. PMID: 35434713 Free PMC article.
-
Structural Characteristics of Heparin Binding to SARS-CoV-2 Spike Protein RBD of Omicron Sub-Lineages BA.2.12.1, BA.4 and BA.5.Viruses. 2022 Dec 1;14(12):2696. doi: 10.3390/v14122696. Viruses. 2022. PMID: 36560700 Free PMC article.
-
Real-Time Analysis of SARS-CoV-2-Induced Cytolysis Reveals Distinct Variant-Specific Replication Profiles.Viruses. 2023 Sep 16;15(9):1937. doi: 10.3390/v15091937. Viruses. 2023. PMID: 37766343 Free PMC article.
References
-
- World Health Organization Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. [(accessed on 12 December 2021)]. Available online: https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.....
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

