Cell Compartment-Specific Folding of Ty1 Long Terminal Repeat Retrotransposon RNA Genome
- PMID: 36146813
- PMCID: PMC9503155
- DOI: 10.3390/v14092007
Cell Compartment-Specific Folding of Ty1 Long Terminal Repeat Retrotransposon RNA Genome
Abstract
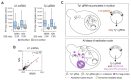
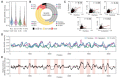
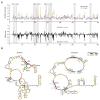
The structural transitions RNAs undergo during trafficking are not well understood. Here, we used the well-developed yeast Ty1 retrotransposon to provide the first structural model of genome (g) RNA in the nucleus from a retrovirus-like transposon. Through a detailed comparison of nuclear Ty1 gRNA structure with those established in the cytoplasm, virus-like particles (VLPs), and those synthesized in vitro, we detected Ty1 gRNA structural alterations that occur during retrotransposition. Full-length Ty1 gRNA serves as the mRNA for Gag and Gag-Pol proteins and as the genome that is reverse transcribed within VLPs. We show that about 60% of base pairs predicted for the nuclear Ty1 gRNA appear in the cytoplasm, and active translation does not account for such structural differences. Most of the shared base pairs are represented by short-range interactions, whereas the long-distance pairings seem unique for each compartment. Highly structured motifs tend to be preserved after nuclear export of Ty1 gRNA. In addition, our study highlights the important role of Ty1 Gag in mediating critical RNA-RNA interactions required for retrotransposition.
Keywords: Gag; LTR-retrotransposon; RNA genome; RNA structure; Ty1; cell compartment-specific folding; gRNA cyclization; gRNA dimerization; tRNA annealing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Retroviral-like determinants and functions required for dimerization of Ty1 retrotransposon RNA.RNA Biol. 2019 Dec;16(12):1749-1763. doi: 10.1080/15476286.2019.1657370. Epub 2019 Aug 30. RNA Biol. 2019. PMID: 31469343 Free PMC article.
-
RNA Binding Properties of the Ty1 LTR-Retrotransposon Gag Protein.Int J Mol Sci. 2021 Aug 23;22(16):9103. doi: 10.3390/ijms22169103. Int J Mol Sci. 2021. PMID: 34445809 Free PMC article.
-
Determinants of Genomic RNA Encapsidation in the Saccharomyces cerevisiae Long Terminal Repeat Retrotransposons Ty1 and Ty3.Viruses. 2016 Jul 14;8(7):193. doi: 10.3390/v8070193. Viruses. 2016. PMID: 27428991 Free PMC article. Review.
-
Ty1 gag enhances the stability and nuclear export of Ty1 mRNA.Traffic. 2013 Jan;14(1):57-69. doi: 10.1111/tra.12013. Epub 2012 Oct 17. Traffic. 2013. PMID: 22998189 Free PMC article.
-
The Ty1 LTR-Retrotransposon of Budding Yeast, Saccharomyces cerevisiae.Microbiol Spectr. 2015 Apr;3(2):MDNA3-0053-2014. doi: 10.1128/microbiolspec.MDNA3-0053-2014. Microbiol Spectr. 2015. PMID: 26104690 Review.
Cited by
-
Mapping the structural landscape of the yeast Ty3 retrotransposon RNA genome.Nucleic Acids Res. 2024 Sep 9;52(16):9821-9837. doi: 10.1093/nar/gkae494. Nucleic Acids Res. 2024. PMID: 38864374 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

