Influenza A (N1-N9) and Influenza B (B/Victoria and B/Yamagata) Neuraminidase Pseudotypes as Tools for Pandemic Preparedness and Improved Influenza Vaccine Design
- PMID: 36146598
- PMCID: PMC9571397
- DOI: 10.3390/vaccines10091520
Influenza A (N1-N9) and Influenza B (B/Victoria and B/Yamagata) Neuraminidase Pseudotypes as Tools for Pandemic Preparedness and Improved Influenza Vaccine Design
Abstract
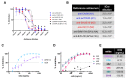
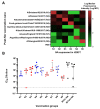
To better understand how inhibition of the influenza neuraminidase (NA) protein contributes to protection against influenza, we produced lentiviral vectors pseudotyped with an avian H11 hemagglutinin (HA) and the NA of all influenza A (N1-N9) subtypes and influenza B (B/Victoria and B/Yamagata). These NA viral pseudotypes (PV) possess stable NA activity and can be utilized as target antigens in in vitro assays to assess vaccine immunogenicity. Employing these NA PV, we developed an enzyme-linked lectin assay (pELLA) for routine serology to measure neuraminidase inhibition (NI) titers of reference antisera, monoclonal antibodies and post-vaccination sera with various influenza antigens. We also show that the pELLA is more sensitive than the commercially available NA-Fluor™ in detecting NA inhibition in these samples. Our studies may lead to establishing the protective NA titer that contributes to NA-based immunity. This will aid in the design of superior, longer lasting and more broadly protective vaccines that can be employed together with HA-targeted vaccines in a pre-pandemic approach.
Keywords: ELLA; antisera; immunity; influenza; inhibition; monoclonal antibody; neuraminidase; pseudotype; vaccine.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Pseudotyped Viruses for Influenza.Adv Exp Med Biol. 2023;1407:153-173. doi: 10.1007/978-981-99-0113-5_8. Adv Exp Med Biol. 2023. PMID: 36920696
-
Validation of a Harmonized Enzyme-Linked-Lectin-Assay (ELLA-NI) Based Neuraminidase Inhibition Assay Standard Operating Procedure (SOP) for Quantification of N1 Influenza Antibodies and the Use of a Calibrator to Improve the Reproducibility of the ELLA-NI With Reverse Genetics Viral and Recombinant Neuraminidase Antigens: A FLUCOP Collaborative Study.Front Immunol. 2022 Jun 17;13:909297. doi: 10.3389/fimmu.2022.909297. eCollection 2022. Front Immunol. 2022. PMID: 35784305 Free PMC article.
-
Universal Influenza Virus Neuraminidase Vaccine Elicits Protective Immune Responses against Human Seasonal and Pre-pandemic Strains.J Virol. 2021 Aug 10;95(17):e0075921. doi: 10.1128/JVI.00759-21. Epub 2021 Aug 10. J Virol. 2021. PMID: 34160258 Free PMC article.
-
Pseudotype-based neutralization assays for influenza: a systematic analysis.Front Immunol. 2015 Apr 29;6:161. doi: 10.3389/fimmu.2015.00161. eCollection 2015. Front Immunol. 2015. PMID: 25972865 Free PMC article. Review.
-
In the shadow of hemagglutinin: a growing interest in influenza viral neuraminidase and its role as a vaccine antigen.Viruses. 2014 Jun 23;6(6):2465-94. doi: 10.3390/v6062465. Viruses. 2014. PMID: 24960271 Free PMC article. Review.
Cited by
-
Conference Report: LPMHealthcare Emerging Viruses 2023 (EVOX23): Pandemics-Learning from the Past and Present to Prepare for the Future.Pathogens. 2024 Aug 10;13(8):679. doi: 10.3390/pathogens13080679. Pathogens. 2024. PMID: 39204279 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

