Rolling Circles as a Means of Encoding Genes in the RNA World
- PMID: 36143408
- PMCID: PMC9505818
- DOI: 10.3390/life12091373
Rolling Circles as a Means of Encoding Genes in the RNA World
Abstract
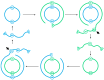
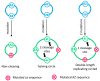
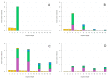
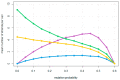
The rolling circle mechanism found in viroids and some RNA viruses is a likely way that replication could have begun in the RNA World. Here, we consider simulations of populations of protocells, each containing multiple copies of rolling circle RNAs that can replicate non-enzymatically. The mechanism requires the presence of short self-cleaving ribozymes such as hammerheads, which can cleave and re-circularize RNA strands. A rolling circle must encode a hammerhead and the complement of a hammerhead, so that both plus and minus strands can cleave. Thus, the minimal functional length is twice the length of the hammerhead sequence. Selection for speed of replication will tend to reduce circles to this minimum length. However, if sequence errors occur when copying the hammerhead sequence, this prevents cleavage at one point, but still allows cleavage on the next passage around the rolling circle. Thus, there is a natural doubling mechanism that creates strands that are multiple times the length of the minimal sequence. This can provide space for the origin of new genes with beneficial functions. We show that if a beneficial gene appears in this new space, the longer sequence with the beneficial function can be selected, even though it replicates more slowly. This provides a route for the evolution of longer circles encoding multiple genes.
Keywords: RNA world; computer simulation; error threshold; rolling circle.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
Similar articles
-
Rolling circle RNA synthesis catalyzed by RNA.Elife. 2022 Feb 2;11:e75186. doi: 10.7554/eLife.75186. Elife. 2022. PMID: 35108196 Free PMC article.
-
Viroid replication: rolling-circles, enzymes and ribozymes.Viruses. 2009 Sep;1(2):317-34. doi: 10.3390/v1020317. Epub 2009 Sep 14. Viruses. 2009. PMID: 21994552 Free PMC article.
-
Rolling-circle and strand-displacement mechanisms for non-enzymatic RNA replication at the time of the origin of life.J Theor Biol. 2021 Oct 21;527:110822. doi: 10.1016/j.jtbi.2021.110822. Epub 2021 Jun 29. J Theor Biol. 2021. PMID: 34214567
-
Viroids: the minimal non-coding RNAs with autonomous replication.FEBS Lett. 2004 Jun 1;567(1):42-8. doi: 10.1016/j.febslet.2004.03.118. FEBS Lett. 2004. PMID: 15165891 Review.
-
Viroids with hammerhead ribozymes: some unique structural and functional aspects with respect to other members of the group.Biol Chem. 1999 Jul-Aug;380(7-8):849-54. doi: 10.1515/BC.1999.104. Biol Chem. 1999. PMID: 10494833 Review.
Cited by
-
The RNA-DNA world and the emergence of DNA-encoded heritable traits.RNA Biol. 2024 Jan;21(1):1-9. doi: 10.1080/15476286.2024.2355391. Epub 2024 May 24. RNA Biol. 2024. PMID: 38785360 Free PMC article.
-
Emergence of catalytic function in prebiotic information-coding polymers.Elife. 2024 Mar 26;12:RP91397. doi: 10.7554/eLife.91397. Elife. 2024. PMID: 38530342 Free PMC article.
-
Circular at the very beginning: on the initial genomes in the RNA world.RNA Biol. 2024 Jan;21(1):17-31. doi: 10.1080/15476286.2024.2380130. Epub 2024 Jul 17. RNA Biol. 2024. PMID: 39016036 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

