Chloroplast Genome Annotation Tools: Prolegomena to the Identification of Inverted Repeats
- PMID: 36142721
- PMCID: PMC9503105
- DOI: 10.3390/ijms231810804
Chloroplast Genome Annotation Tools: Prolegomena to the Identification of Inverted Repeats
Abstract
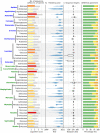
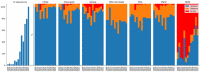
The development of next-generation sequencing technology and the increasing amount of sequencing data have brought the bioinformatic tools used in genome assembly into focus. The final step of the process is genome annotation, which works on assembled genome sequences to identify the location of genome features. In the case of organelle genomes, specialized annotation tools are used to identify organelle genes and structural features. Numerous annotation tools target chloroplast sequences. Most chloroplast DNA genomes have a quadripartite structure caused by two copies of a large inverted repeat. We investigated the strategies of six annotation tools (Chloë, Chloroplot, GeSeq, ORG.Annotate, PGA, Plann) for identifying inverted repeats and analyzed their success using publicly available complete chloroplast sequences of taxa belonging to the asterid and rosid clades. The annotation tools use two different approaches to identify inverted repeats, using existing general search tools or implementing stand-alone solutions. The chloroplast sequences studied show that there are different types of imperfections in the assembled data and that each tool performs better on some sequences than the others.
Keywords: annotation; chloroplast genome; inverted repeats; repeat identification.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Comparison of different annotation tools for characterization of the complete chloroplast genome of Corylus avellana cv Tombul.BMC Genomics. 2019 Nov 20;20(1):874. doi: 10.1186/s12864-019-6253-5. BMC Genomics. 2019. PMID: 31747873 Free PMC article.
-
Complete Chloroplast Genome Sequences of Kaempferia Galanga and Kaempferia Elegans: Molecular Structures and Comparative Analysis.Molecules. 2019 Jan 29;24(3):474. doi: 10.3390/molecules24030474. Molecules. 2019. PMID: 30699955 Free PMC article.
-
Phylogenetic studies and comparative chloroplast genome analyses elucidate the basal position of halophyte Nitraria sibirica (Nitrariaceae) in the Sapindales.Mitochondrial DNA A DNA Mapp Seq Anal. 2018 Jul;29(5):745-755. doi: 10.1080/24701394.2017.1350954. Epub 2017 Jul 15. Mitochondrial DNA A DNA Mapp Seq Anal. 2018. PMID: 28712318
-
Activity-based annotation: the emergence of systems biochemistry.Trends Biochem Sci. 2022 Sep;47(9):785-794. doi: 10.1016/j.tibs.2022.03.017. Epub 2022 Apr 13. Trends Biochem Sci. 2022. PMID: 35430135 Free PMC article. Review.
-
Diversity and features of proteins with structural repeats.Biophys Rev. 2023 Sep 7;15(5):1159-1169. doi: 10.1007/s12551-023-01130-0. eCollection 2023 Oct. Biophys Rev. 2023. PMID: 37974986 Free PMC article. Review.
Cited by
-
Analysis of the Chloroplast Genome of Ficus simplicissima Lour Collected in Vietnam and Proposed Barcodes for Identifying Ficus Plants.Curr Issues Mol Biol. 2023 Jan 27;45(2):1024-1036. doi: 10.3390/cimb45020067. Curr Issues Mol Biol. 2023. PMID: 36826012 Free PMC article.
-
Progress, challenge and prospect of plant plastome annotation.Front Plant Sci. 2023 May 30;14:1166140. doi: 10.3389/fpls.2023.1166140. eCollection 2023. Front Plant Sci. 2023. PMID: 37324662 Free PMC article. Review.
-
CPJSdraw: analysis and visualization of junction sites of chloroplast genomes.PeerJ. 2023 May 10;11:e15326. doi: 10.7717/peerj.15326. eCollection 2023. PeerJ. 2023. PMID: 37193025 Free PMC article.
-
Variation in Chloroplast Genome Size: Biological Phenomena and Technological Artifacts.Plants (Basel). 2023 Jan 5;12(2):254. doi: 10.3390/plants12020254. Plants (Basel). 2023. PMID: 36678967 Free PMC article.
-
Assembly, annotation and analysis of the chloroplast genome of the Algarrobo tree Neltuma pallida (subfamily: Caesalpinioideae).BMC Plant Biol. 2023 Nov 16;23(1):570. doi: 10.1186/s12870-023-04581-5. BMC Plant Biol. 2023. PMID: 37974117 Free PMC article.
References
-
- Dagan T., Roettger M., Stucken K., Landan G., Koch R., Major P., Gould S.B., Goremykin V.V., Rippka R., De Marsac N.T., et al. Genomes of Stigonematalean Cyanobacteria (Subsection V) and the Evolution of Oxygenic Photosynthesis from Prokaryotes to Plastids. Genome Biol. Evol. 2013;5:31–44. doi: 10.1093/gbe/evs117. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

