New Inhibitors of the Human p300/CBP Acetyltransferase Are Selectively Active against the Arabidopsis HAC Proteins
- PMID: 36142359
- PMCID: PMC9499386
- DOI: 10.3390/ijms231810446
New Inhibitors of the Human p300/CBP Acetyltransferase Are Selectively Active against the Arabidopsis HAC Proteins
Abstract
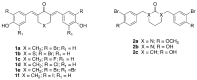
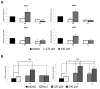
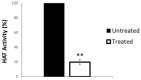
Histone acetyltransferases (HATs) are involved in the epigenetic positive control of gene expression in eukaryotes. CREB-binding proteins (CBP)/p300, a subfamily of highly conserved HATs, have been shown to function as acetylases on both histones and non-histone proteins. In the model plant Arabidopsis thaliana among the five CBP/p300 HATs, HAC1, HAC5 and HAC12 have been shown to be involved in the ethylene signaling pathway. In addition, HAC1 and HAC5 interact and cooperate with the Mediator complex, as in humans. Therefore, it is potentially difficult to discriminate the effect on plant development of the enzymatic activity with respect to their Mediator-related function. Taking advantage of the homology of the human HAC catalytic domain with that of the Arabidopsis, we set-up a phenotypic assay based on the hypocotyl length of Arabidopsis dark-grown seedlings to evaluate the effects of a compound previously described as human p300/CBP inhibitor, and to screen previously described cinnamoyl derivatives as well as newly synthesized analogues. We selected the most effective compounds, and we demonstrated their efficacy at phenotypic and molecular level. The in vitro inhibition of the enzymatic activity proved the specificity of the inhibitor on the catalytic domain of HAC1, thus substantiating this strategy as a useful tool in plant epigenetic studies.
Keywords: Arabidopsis thaliana; HAC proteins; p300/CBP inhibitors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
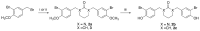


Similar articles
-
The CBP/p300 histone acetyltransferases function as plant-specific MEDIATOR subunits in Arabidopsis.J Integr Plant Biol. 2021 Apr;63(4):755-771. doi: 10.1111/jipb.13052. Epub 2021 Mar 8. J Integr Plant Biol. 2021. PMID: 33325122
-
Involvement of Arabidopsis HAC family genes in pleiotropic developmental processes.Plant Signal Behav. 2014;9(3):e28173. doi: 10.4161/psb.28173. Epub 2014 Mar 10. Plant Signal Behav. 2014. PMID: 24614176 Free PMC article.
-
Involvement of Arabidopsis histone acetyltransferase HAC family genes in the ethylene signaling pathway.Plant Cell Physiol. 2014 Feb;55(2):426-35. doi: 10.1093/pcp/pct180. Epub 2013 Nov 28. Plant Cell Physiol. 2014. PMID: 24287137
-
Structure and chemistry of the p300/CBP and Rtt109 histone acetyltransferases: implications for histone acetyltransferase evolution and function.Curr Opin Struct Biol. 2008 Dec;18(6):741-7. doi: 10.1016/j.sbi.2008.09.004. Epub 2008 Oct 27. Curr Opin Struct Biol. 2008. PMID: 18845255 Free PMC article. Review.
-
Recent Advances in the Development of CBP/p300 Bromodomain Inhibitors.Curr Med Chem. 2020;27(33):5583-5598. doi: 10.2174/0929867326666190731141055. Curr Med Chem. 2020. PMID: 31364510 Review.
Cited by
-
Regulatory Mechanisms of Transcription Factors in Plant Morphology and Function.Int J Mol Sci. 2023 Apr 11;24(8):7039. doi: 10.3390/ijms24087039. Int J Mol Sci. 2023. PMID: 37108201 Free PMC article.
-
Plants and Small Molecules: An Up-and-Coming Synergy.Plants (Basel). 2023 Apr 21;12(8):1729. doi: 10.3390/plants12081729. Plants (Basel). 2023. PMID: 37111951 Free PMC article. Review.
-
Genome-wide identification of Brassicaceae histone modification genes and their responses to abiotic stresses in allotetraploid rapeseed.BMC Plant Biol. 2023 May 11;23(1):248. doi: 10.1186/s12870-023-04256-1. BMC Plant Biol. 2023. PMID: 37170202 Free PMC article.
-
How Histone Acetyltransferases Shape Plant Photomorphogenesis and UV Response.Int J Mol Sci. 2024 Jul 18;25(14):7851. doi: 10.3390/ijms25147851. Int J Mol Sci. 2024. PMID: 39063093 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

