SR Splicing Factors Promote Cancer via Multiple Regulatory Mechanisms
- PMID: 36140826
- PMCID: PMC9498594
- DOI: 10.3390/genes13091659
SR Splicing Factors Promote Cancer via Multiple Regulatory Mechanisms
Abstract
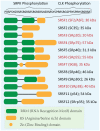
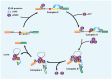
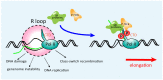
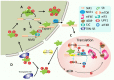
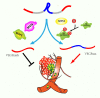
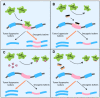
Substantial emerging evidence supports that dysregulated RNA metabolism is associated with tumor initiation and development. Serine/Arginine-Rich proteins (SR) are a number of ultraconserved and structurally related proteins that contain a characteristic RS domain rich in arginine and serine residues. SR proteins perform a critical role in spliceosome assembling and conformational transformation, contributing to precise alternative RNA splicing. Moreover, SR proteins have been reported to participate in multiple other RNA-processing-related mechanisms than RNA splicing, such as genome stability, RNA export, and translation. The dysregulation of SR proteins has been reported to contribute to tumorigenesis through multiple mechanisms. Here we reviewed the different biological roles of SR proteins and strategies for functional rectification of SR proteins that may serve as potential therapeutic approaches for cancer.
Keywords: RNA processing; SR proteins; cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Comment in
-
RNA Splicing in Cancer and Targeted Therapies.Genes (Basel). 2023 Oct 29;14(11):2020. doi: 10.3390/genes14112020. Genes (Basel). 2023. PMID: 38002963 Free PMC article.
Similar articles
-
SR proteins in cancer: function, regulation, and small inhibitor.Cell Mol Biol Lett. 2024 May 22;29(1):78. doi: 10.1186/s11658-024-00594-6. Cell Mol Biol Lett. 2024. PMID: 38778254 Free PMC article. Review.
-
Targeting serine- and arginine-rich splicing factors to rectify aberrant alternative splicing.Drug Discov Today. 2023 Sep;28(9):103691. doi: 10.1016/j.drudis.2023.103691. Epub 2023 Jun 27. Drug Discov Today. 2023. PMID: 37385370 Review.
-
Regulatory Network of Serine/Arginine-Rich (SR) Proteins: The Molecular Mechanism and Physiological Function in Plants.Int J Mol Sci. 2022 Sep 5;23(17):10147. doi: 10.3390/ijms231710147. Int J Mol Sci. 2022. PMID: 36077545 Free PMC article. Review.
-
SR Proteins: Binders, Regulators, and Connectors of RNA.Mol Cells. 2017 Jan;40(1):1-9. doi: 10.14348/molcells.2017.2319. Epub 2017 Jan 26. Mol Cells. 2017. PMID: 28152302 Free PMC article. Review.
-
A critical update on the strategies towards small molecule inhibitors targeting Serine/arginine-rich (SR) proteins and Serine/arginine-rich proteins related kinases in alternative splicing.Bioorg Med Chem. 2022 Sep 15;70:116921. doi: 10.1016/j.bmc.2022.116921. Epub 2022 Jul 9. Bioorg Med Chem. 2022. PMID: 35863237 Review.
Cited by
-
Long non-coding RNA SNHG9 regulates viral replication in rhabdomyosarcoma cells infected with enterovirus D68 via miR-150-5p/c-Fos axis.Front Microbiol. 2023 Jan 19;13:1081237. doi: 10.3389/fmicb.2022.1081237. eCollection 2022. Front Microbiol. 2023. PMID: 36741904 Free PMC article.
-
DNA Repair and Therapeutic Strategies in Cancer Stem Cells.Cancers (Basel). 2023 Mar 22;15(6):1897. doi: 10.3390/cancers15061897. Cancers (Basel). 2023. PMID: 36980782 Free PMC article. Review.
-
High expression of SRSF1 facilitates osteosarcoma progression and unveils its potential mechanisms.BMC Cancer. 2024 May 12;24(1):580. doi: 10.1186/s12885-024-12346-y. BMC Cancer. 2024. PMID: 38735973 Free PMC article.
-
Study of prognostic splicing factors in cancer using machine learning approaches.Hum Mol Genet. 2024 Jun 21;33(13):1131-1141. doi: 10.1093/hmg/ddae047. Hum Mol Genet. 2024. PMID: 38538560
-
SR proteins in cancer: function, regulation, and small inhibitor.Cell Mol Biol Lett. 2024 May 22;29(1):78. doi: 10.1186/s11658-024-00594-6. Cell Mol Biol Lett. 2024. PMID: 38778254 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

