Folding and self-assembly of short intrinsically disordered peptides and protein regions
- PMID: 36133101
- PMCID: PMC9417027
- DOI: 10.1039/d0na00941e
Folding and self-assembly of short intrinsically disordered peptides and protein regions
Abstract
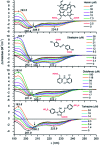
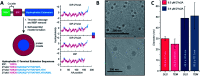
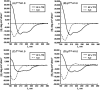
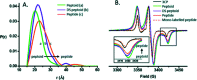
Proteins and peptide fragments are highly relevant building blocks in self-assembly for nanostructures with plenty of applications. Intrinsically disordered proteins (IDPs) and protein regions (IDRs) are defined by the absence of a well-defined secondary structure, yet IDPs/IDRs show a significant biological activity. Experimental techniques and computational modelling procedures for the characterization of IDPs/IDRs are discussed. Directed self-assembly of IDPs/IDRs allows reaching a large variety of nanostructures. Hybrid materials based on the derivatives of IDPs/IDRs show a promising performance as alternative biocides and nanodrugs. Cell mimicking, in vivo compartmentalization, and bone regeneration are demonstrated for IDPs/IDRs in biotechnological applications. The exciting possibilities of IDPs/IDRs in nanotechnology with relevant biological applications are shown.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures



Similar articles
-
Features of molecular recognition of intrinsically disordered proteins via coupled folding and binding.Protein Sci. 2019 Nov;28(11):1952-1965. doi: 10.1002/pro.3718. Epub 2019 Sep 4. Protein Sci. 2019. PMID: 31441158 Free PMC article. Review.
-
Computational Prediction of Protein Intrinsically Disordered Region Related Interactions and Functions.Genes (Basel). 2023 Feb 8;14(2):432. doi: 10.3390/genes14020432. Genes (Basel). 2023. PMID: 36833360 Free PMC article. Review.
-
Unfoldomics of human diseases: linking protein intrinsic disorder with diseases.BMC Genomics. 2009 Jul 7;10 Suppl 1(Suppl 1):S7. doi: 10.1186/1471-2164-10-S1-S7. BMC Genomics. 2009. PMID: 19594884 Free PMC article.
-
Do sequence neighbours of intrinsically disordered regions promote structural flexibility in intrinsically disordered proteins?J Struct Biol. 2020 Feb 1;209(2):107428. doi: 10.1016/j.jsb.2019.107428. Epub 2019 Nov 20. J Struct Biol. 2020. PMID: 31756456
-
Conservation and coevolution determine evolvability of different classes of disordered residues in human intrinsically disordered proteins.Proteins. 2022 Mar;90(3):632-644. doi: 10.1002/prot.26261. Epub 2021 Oct 14. Proteins. 2022. PMID: 34626492
Cited by
-
A Molecular Mechanism to Explain the Nickel-Induced Changes in Protamine-like Proteins and Their DNA Binding Affecting Sperm Chromatin in Mytilus galloprovincialis: An In Vitro Study.Biomolecules. 2023 Mar 12;13(3):520. doi: 10.3390/biom13030520. Biomolecules. 2023. PMID: 36979455 Free PMC article.
-
Alternative Causal Link between Peptide Fibrillization and β-Strand Conformation.ACS Omega. 2021 May 5;6(19):12904-12912. doi: 10.1021/acsomega.1c01423. eCollection 2021 May 18. ACS Omega. 2021. PMID: 34056442 Free PMC article.
-
Self-Assembling Peptides: From Design to Biomedical Applications.Int J Mol Sci. 2021 Nov 23;22(23):12662. doi: 10.3390/ijms222312662. Int J Mol Sci. 2021. PMID: 34884467 Free PMC article. Review.
-
From isolated polyelectrolytes to star-like assemblies: the role of sequence heterogeneity on the statistical structure of the intrinsically disordered neurofilament-low tail domain.Eur Phys J E Soft Matter. 2024 Feb 15;47(2):13. doi: 10.1140/epje/s10189-024-00409-8. Eur Phys J E Soft Matter. 2024. PMID: 38358563 Free PMC article.
-
How well does molecular simulation reproduce environment-specific conformations of the intrinsically disordered peptides PLP, TP2 and ONEG?Chem Sci. 2022 Jan 20;13(7):1957-1971. doi: 10.1039/d1sc03496k. eCollection 2022 Feb 16. Chem Sci. 2022. PMID: 35308859 Free PMC article.
References
-
- Li S. Xing R. Chang R. Zou Q. Yan X. Curr. Opin. Colloid Interface Sci. 2018;35:17–25. doi: 10.1016/j.cocis.2017.12.004. - DOI
Publication types
LinkOut - more resources
Full Text Sources

