VAMP724 and VAMP726 are involved in autophagosome formation in Arabidopsis thaliana
- PMID: 36130166
- PMCID: PMC10240985
- DOI: 10.1080/15548627.2022.2127240
VAMP724 and VAMP726 are involved in autophagosome formation in Arabidopsis thaliana
Abstract
Macroautophagy/autophagy, an evolutionarily conserved degradative process essential for cell homeostasis and development in eukaryotes, involves autophagosome formation and fusion with a lysosome/vacuole. The soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins play important roles in regulating autophagy in mammals and yeast, but relatively little is known about SNARE function in plant autophagy. Here we identified and characterized two Arabidopsis SNAREs, AT4G15780/VAMP724 and AT1G04760/VAMP726, involved in plant autophagy. Phenotypic analysis showed that mutants of VAMP724 and VAMP726 are sensitive to nutrient-starved conditions. Live-cell imaging on mutants of VAMP724 and VAMP726 expressing YFP-ATG8e showed the formation of abnormal autophagic structures outside the vacuoles and compromised autophagic flux. Further immunogold transmission electron microscopy and electron tomography (ET) analysis demonstrated a direct connection between the tubular autophagic structures and the endoplasmic reticulum (ER) in vamp724-1 vamp726-1 double mutants. Further transient co-expression, co-immunoprecipitation and double immunogold TEM analysis showed that ATG9 (autophagy related 9) interacts and colocalizes with VAMP724 and VAMP726 in ATG9-positive vesicles during autophagosome formation. Taken together, VAMP724 and VAMP726 regulate autophagosome formation likely working together with ATG9 in Arabidopsis.Abbreviations: ATG, autophagy related; BTH, benzo-(1,2,3)-thiadiazole-7-carbothioic acid S-methyl ester; Conc A, concanamycin A; EM, electron microscopy; ER, endoplasmic reticulum; FRET, Förster/fluorescence resonance energy transfer; MS, Murashige and Skoog; MVB, multivesicular body; PAS, phagophore assembly site; PM, plasma membrane; PVC, prevacuolar compartment; SNARE, soluble N-ethylmaleimide-sensitive factor attachment protein receptor; TEM, transmission electron microscopy; TGN, trans-Golgi network; WT, wild-type.
Keywords: ATG9; Arabidopsis; SNARE; VAMP724; VAMP726; autophagy.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
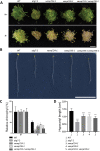
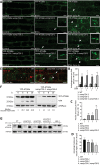
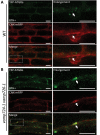

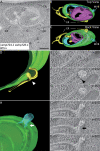

Similar articles
-
ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis.Proc Natl Acad Sci U S A. 2017 Jan 17;114(3):E426-E435. doi: 10.1073/pnas.1616299114. Epub 2017 Jan 4. Proc Natl Acad Sci U S A. 2017. PMID: 28053229 Free PMC article.
-
A nuclear membrane-derived structure associated with Atg8 is involved in the sequestration of selective cargo, the Cvt complex, during autophagosome formation in yeast.Autophagy. 2019 Mar;15(3):423-437. doi: 10.1080/15548627.2018.1525475. Epub 2018 Oct 11. Autophagy. 2019. PMID: 30238844 Free PMC article.
-
Arabidopsis AUTOPHAGY-RELATED2 is essential for ATG18a and ATG9 trafficking during autophagosome closure.Plant Physiol. 2023 Aug 31;193(1):304-321. doi: 10.1093/plphys/kiad287. Plant Physiol. 2023. PMID: 37195145
-
New insights regarding SNARE proteins in autophagosome-lysosome fusion.Autophagy. 2021 Oct;17(10):2680-2688. doi: 10.1080/15548627.2020.1823124. Epub 2020 Sep 24. Autophagy. 2021. PMID: 32924745 Free PMC article. Review.
-
The Atg17-Atg31-Atg29 complex and Atg11 regulate autophagosome-vacuole fusion.Autophagy. 2016 May 3;12(5):894-5. doi: 10.1080/15548627.2016.1162364. Autophagy. 2016. PMID: 26986547 Free PMC article. Review.
Cited by
-
Dynamic organelle changes and autophagic processes in lily pollen germination.Bot Stud. 2024 Jan 26;65(1):5. doi: 10.1186/s40529-024-00410-6. Bot Stud. 2024. PMID: 38273136 Free PMC article.
-
Autophagosome development and chloroplast segmentation occur synchronously for piecemeal degradation of chloroplasts.Elife. 2024 Nov 7;12:RP93232. doi: 10.7554/eLife.93232. Elife. 2024. PMID: 39509463 Free PMC article.
-
VAMP726 from maize and Arabidopsis confers pollen resistance to heat and UV radiation by influencing lignin content of sporopollenin.Plant Commun. 2023 Nov 13;4(6):100682. doi: 10.1016/j.xplc.2023.100682. Epub 2023 Sep 9. Plant Commun. 2023. PMID: 37691288 Free PMC article.
-
Autophagosome biogenesis and organelle homeostasis in plant cells.Plant Cell. 2024 Sep 3;36(9):3009-3024. doi: 10.1093/plcell/koae099. Plant Cell. 2024. PMID: 38536783 Review.
-
VAMP726 and VAMP725 regulate vesicle secretion and pollen tube growth in Arabidopsis.Plant Cell Rep. 2023 Dec;42(12):1951-1965. doi: 10.1007/s00299-023-03075-w. Epub 2023 Oct 8. Plant Cell Rep. 2023. PMID: 37805949
References
-
- Xie Z, Klionsky DJ.. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9(10):1102–9. - PubMed
-
- Liu Y, Bassham DC.. Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol. 2012;63:215–37. - PubMed
-
- Morishita H, Mizushima N.. Diverse cellular roles of autophagy. Annu Rev Cell Dev Biol. 2019;35:453–475. - PubMed
-
- Lipka V, Kwon C, Panstruga R.. SNARE-ware: the role of SNARE-domain proteins in plant biology. Annu Rev Cell Dev Biol. 2007;23:147–74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
