Fine mapping a quantitative trait locus underlying seedling resistance to gummy stem blight using a residual heterozygous lines-derived strategy in cucumber
- PMID: 36119620
- PMCID: PMC9480501
- DOI: 10.3389/fpls.2022.968811
Fine mapping a quantitative trait locus underlying seedling resistance to gummy stem blight using a residual heterozygous lines-derived strategy in cucumber
Abstract
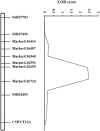
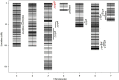
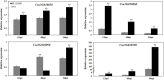
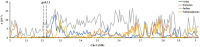
Gummy stem blight (GSB), caused by Didymella bryoniae, is one of the most devastating diseases that severely reduces cucumber production. Developing resistant varieties would be an effective strategy to control GSB. Although several GSB-resistant QTLs have been reported, causal genes for GSB resistance have not yet been identified in cucumber. A novel loci gsb3.1 for seedling GSB resistance from the "PI 183967" genotype was previously identified in a 1.7-Mb interval on chromosome 3. In this study, we developed a residual heterozygous line-derived strategy from Recombinant Inbred Lines to perform fine mapping, and with this approach, the gsb3.1 locus was narrowed to a 38 kb interval. There were six predicted genes at the gsb3.1 locus, four of which differed in expression in the GSB-resistant compared to the susceptible lines after fungal inoculation. These candidate genes (Csa3G020050, Csa3G020060, Csa3G020090, and Csa3G020590) within the gsb3.1 locus could be helpful for the genetic study of GSB resistance and marker-assisted selection in cucumber. Phylogenetic analyses indicated that the resistant gsb3.1 allele may uniquely exist in the wild species present in the Indian group, and that nucleotide diversity was significantly reduced in cultivated accessions. Therefore, the gsb3.1 allele could be introgressed into existing commercial cultivars and combined with other resistance QTLs to provide broad-spectrum and robust GSB resistance in cucumber.
Keywords: Cucumis sativus L.; candidate genes; fine-mapping; gummy stem blight; residual heterozygous line (RHL)-derived strategy.
Copyright © 2022 Han, Dong, Liu, Shi, Beckles, Gu, Miao and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Gummy stem blight: One disease, three pathogens.Mol Plant Pathol. 2023 Aug;24(8):825-837. doi: 10.1111/mpp.13339. Epub 2023 May 2. Mol Plant Pathol. 2023. PMID: 37129449 Free PMC article. Review.
-
Genetic Analysis and QTL Mapping of Resistance to Gummy Stem Blight in Cucumis sativus Seedling Stage.Plant Dis. 2017 Jul;101(7):1145-1152. doi: 10.1094/PDIS-08-16-1116-RE. Epub 2017 Apr 27. Plant Dis. 2017. PMID: 30682960
-
Fine mapping and candidate gene analysis of gummy stem blight resistance in cucumber stem.Theor Appl Genet. 2022 Sep;135(9):3117-3125. doi: 10.1007/s00122-022-04172-2. Epub 2022 Jul 23. Theor Appl Genet. 2022. PMID: 35869997
-
Identification and Molecular Mapping of a Gummy Stem Blight Resistance Gene in Wild Watermelon (Citrullus amarus) Germplasm PI 189225.Plant Dis. 2020 Jan;104(1):16-24. doi: 10.1094/PDIS-04-19-0753-RE. Epub 2019 Nov 15. Plant Dis. 2020. PMID: 31730411
-
Recent status of Genotyping by Sequencing (GBS) Technology in cucumber (Cucumis sativus L.): a review.Mol Biol Rep. 2022 Jun;49(6):5547-5554. doi: 10.1007/s11033-022-07469-z. Epub 2022 May 20. Mol Biol Rep. 2022. PMID: 35596053 Review.
Cited by
-
Gummy stem blight: One disease, three pathogens.Mol Plant Pathol. 2023 Aug;24(8):825-837. doi: 10.1111/mpp.13339. Epub 2023 May 2. Mol Plant Pathol. 2023. PMID: 37129449 Free PMC article. Review.
References
-
- Breitenbach H. H., Wenig M., Wittek F., Jorda L., Maldonado-Alconada A. M., Sarioglu H., et al. . (2014). Contrasting roles of the APOPLASTIC aspartyl protease Apoplastic, Enhanced Disease Susceptibility1-Dependent1 And Legume Lectin-Like Protein1 in Arabidopsis systemic acquired resistance. Plant Physiol. 165, 791–809. doi: 10.1104/pp.114.239665, PMID: - DOI - PMC - PubMed
-
- Chen J., Isshiki S., Tashiro Y., Takeshita A., Miyazaki S. (1995). Studies on a wild cucumber from China. I. Genetic distances between C. hystrix and two cultivated Cucumis species. J. Japan Soc. Hort. Sci. 64, 264–265.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

