Codon Usage Bias and Cluster Analysis of the MMP-2 and MMP-9 Genes in Seven Mammals
- PMID: 36118275
- PMCID: PMC9467794
- DOI: 10.1155/2022/2823356
Codon Usage Bias and Cluster Analysis of the MMP-2 and MMP-9 Genes in Seven Mammals
Abstract
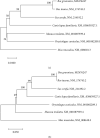
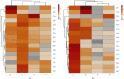
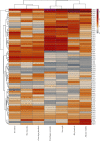
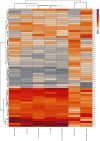
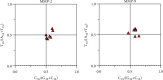
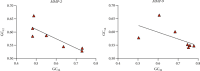
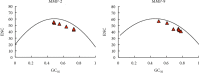
Matrix metalloproteinase (MMP)-2 and MMP-9 are a family of Zn2+ and Ca2+-dependent gelatinase MMPs that regulate muscle development and disease treatment, and they are highly conservative during biological evolution. Despite increasing knowledge of MMP genes, their evolutionary mechanism for functional adaption remains unclear. Moreover, analysis of codon usage bias (CUB) is reliable to understand evolutionary associations. However, the distribution of CUB of MMP-2 and MMP-9 genes in mammals has not been revealed clearly. Multiple analytical software was used to study the genetic evolution, phylogeny, and codon usage pattern of these two genes in seven species of mammals. Results showed that the MMP-2 and MMP-9 genes have CUB. By comparing the content of synonymous codon bases amongst seven mammals, we found that MMP-2 and MMP-9 were low-expression genes in mammals with high codon conservation, and their third codon preferred the G/C base. RSCU analysis revealed that these two genes preferred codons encoding delicious amino acids. Analysing what factors influence CUB showed that the third base distributors of these two genes were C/A and C/T, and GC3S had a wide distribution range on the ENC plot reference curve under no selection or mutational pressure. Thus, mutational pressure is an important factor in CUB. This study revealed the usage characteristics of the MMP-2 and MMP-9 gene codons in different mammals and provided basic data for further study towards enhancing meat flavour, treating muscle disease, and optimizing codons.
Copyright © 2022 Tanliang Ouyang et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures
Similar articles
-
Codon usage characterization and phylogenetic analysis of the mitochondrial genome in Hemerocallis citrina.BMC Genom Data. 2024 Jan 13;25(1):6. doi: 10.1186/s12863-024-01191-4. BMC Genom Data. 2024. PMID: 38218810 Free PMC article.
-
Evolution of Codon Usage Bias in Diatoms.Genes (Basel). 2019 Nov 6;10(11):894. doi: 10.3390/genes10110894. Genes (Basel). 2019. PMID: 31698749 Free PMC article.
-
Influencing elements of codon usage bias in Birnaviridae and its evolutionary analysis.Virus Res. 2022 Mar;310:198672. doi: 10.1016/j.virusres.2021.198672. Epub 2022 Jan 2. Virus Res. 2022. PMID: 34986367
-
Composition, codon usage pattern, protein properties, and influencing factors in the genomes of members of the family Anelloviridae.Arch Virol. 2021 Feb;166(2):461-474. doi: 10.1007/s00705-020-04890-2. Epub 2021 Jan 3. Arch Virol. 2021. PMID: 33392821 Free PMC article.
-
Calculating and comparing codon usage values in rare disease genes highlights codon clustering with disease-and tissue- specific hierarchy.PLoS One. 2022 Mar 31;17(3):e0265469. doi: 10.1371/journal.pone.0265469. eCollection 2022. PLoS One. 2022. PMID: 35358230 Free PMC article.
Cited by
-
Codon Bias of the DDR1 Gene and Transcription Factor EHF in Multiple Species.Int J Mol Sci. 2024 Oct 4;25(19):10696. doi: 10.3390/ijms251910696. Int J Mol Sci. 2024. PMID: 39409024 Free PMC article.
References
-
- Ikemura T. Correlation between the abundance of escherichia coli transfer rnas and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. Journal of Molecular Biology . 1981;151(3):389–409. doi: 10.1016/0022-2836(81)90003-6. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

