Genotypic Resistance Testing of HIV-1 DNA in Peripheral Blood Mononuclear Cells
- PMID: 36102816
- PMCID: PMC9769561
- DOI: 10.1128/cmr.00052-22
Genotypic Resistance Testing of HIV-1 DNA in Peripheral Blood Mononuclear Cells
Abstract
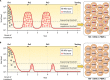
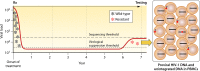
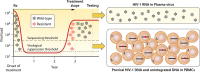
HIV-1 DNA exists in nonintegrated linear and circular episomal forms and as integrated proviruses. In patients with plasma viremia, most peripheral blood mononuclear cell (PBMC) HIV-1 DNA consists of recently produced nonintegrated virus DNA while in patients with prolonged virological suppression (VS) on antiretroviral therapy (ART), most PBMC HIV-1 DNA consists of proviral DNA produced months to years earlier. Drug-resistance mutations (DRMs) in PBMCs are more likely to coexist with ancestral wild-type virus populations than they are in plasma, explaining why next-generation sequencing is particularly useful for the detection of PBMC-associated DRMs. In patients with ongoing high levels of active virus replication, the DRMs detected in PBMCs and in plasma are usually highly concordant. However, in patients with lower levels of virus replication, it may take several months for plasma virus DRMs to reach detectable levels in PBMCs. This time lag explains why, in patients with VS, PBMC genotypic resistance testing (GRT) is less sensitive than historical plasma virus GRT, if previous episodes of virological failure and emergent DRMs were either not prolonged or not associated with high levels of plasma viremia. Despite the increasing use of PBMC GRT in patients with VS, few studies have examined the predictive value of DRMs on the response to a simplified ART regimen. In this review, we summarize what is known about PBMC HIV-1 DNA dynamics, particularly in patients with suppressed plasma viremia, the methods used for PBMC HIV-1 GRT, and the scenarios in which PBMC GRT has been used clinically.
Keywords: DNA sequencing; HIV-1; adaptive mutations; antiviral therapy; drug resistance evolution; peripheral blood mononuclear cells; provirus.
Conflict of interest statement
The authors declare a conflict of interest. Robert W. Shafer has served on two advisory boards for Gilead Sciences and for GlaxoSmithKline Maria Mercedes Santoro has received funds for attending symposia, speaking and organizing educational activities from ViiV Healthcare, Janssen-Cilag and Theratechnologies. Charles Walworth is an Employee, Laboratory Corporation of America, officer of the corporation and shareholder.
Figures
Similar articles
-
Genotypic resistance test in proviral DNA can identify resistance mutations never detected in historical genotypic test in patients with low level or undetectable HIV-RNA.J Clin Virol. 2016 Sep;82:94-100. doi: 10.1016/j.jcv.2016.07.007. Epub 2016 Jul 18. J Clin Virol. 2016. PMID: 27472519
-
Resistance detected in PBMCs predicts virological rebound in HIV-1 suppressed patients switching treatment.J Clin Virol. 2018 Jul;104:61-64. doi: 10.1016/j.jcv.2018.04.001. Epub 2018 Apr 3. J Clin Virol. 2018. PMID: 29738896
-
New Aspects of the Virus Life Cycle and Clinical Utility of Next Generation Sequencing based HIV-1 Resistance Testing in the Genomic, the Proviral, and the Viral Reservoir of Peripheral Blood Mononuclear Cells.Curr HIV Res. 2022;20(3):213-221. doi: 10.2174/1570162X20666220324111418. Curr HIV Res. 2022. PMID: 35331114
-
Molecular biological assessment methods and understanding the course of the HIV infection.APMIS Suppl. 2003;(114):1-37. APMIS Suppl. 2003. PMID: 14626050 Review.
-
Study of the impact of HIV genotypic drug resistance testing on therapy efficacy.Verh K Acad Geneeskd Belg. 2001;63(5):447-73. Verh K Acad Geneeskd Belg. 2001. PMID: 11813503 Review.
Cited by
-
Prevalence of HIV-1 drug resistance mutations in proviral DNA in the Swiss HIV Cohort Study, a retrospective study from 1995 to 2018.J Antimicrob Chemother. 2023 Sep 5;78(9):2323-2334. doi: 10.1093/jac/dkad240. J Antimicrob Chemother. 2023. PMID: 37545164 Free PMC article.
-
Assessing the Virologic Impact of Archived Resistance in the Dolutegravir/Lamivudine 2-Drug Regimen HIV-1 Switch Study TANGO through Week 144.Viruses. 2023 Jun 11;15(6):1350. doi: 10.3390/v15061350. Viruses. 2023. PMID: 37376649 Free PMC article.
-
Dolutegravir/Lamivudine Efficacy and Safety Outcomes in People With HIV-1 With or Without Historical Resistance Results at Screening: 48-Week Pooled Analysis.Open Forum Infect Dis. 2024 Jul 1;11(7):ofae365. doi: 10.1093/ofid/ofae365. eCollection 2024 Jul. Open Forum Infect Dis. 2024. PMID: 39015350 Free PMC article. Clinical Trial.
-
Management of low-level HIV viremia during antiretroviral therapy: Delphi consensus statement and appraisal of the evidence.Sex Transm Infect. 2024 Oct 17;100(7):442-449. doi: 10.1136/sextrans-2024-056199. Sex Transm Infect. 2024. PMID: 39288982 Free PMC article.
-
Practical updates in clinical antiviral resistance testing.J Clin Microbiol. 2024 Aug 14;62(8):e0072823. doi: 10.1128/jcm.00728-23. Epub 2024 Jul 25. J Clin Microbiol. 2024. PMID: 39051778 Free PMC article. Review.
References
-
- DeGruttola V, Dix L, D'Aquila R, Holder D, Phillips A, Ait-Khaled M, Baxter J, Clevenbergh P, Hammer S, Harrigan R, Katzenstein D, Lanier R, Miller M, Para M, Yerly S, Zolopa A, Murray J, Patick A, Miller V, Castillo S, Pedneault L, Mellors J. 2000. The relation between baseline HIV drug resistance and response to antiretroviral therapy: re-analysis of retrospective and prospective studies using a standardized data analysis plan. Antivir Ther 5:41–48. 10.1177/135965350000500112. - DOI - PubMed
-
- Lanier ER, Ait-Khaled M, Scott J, Stone C, Melby T, Sturge G, St Clair M, Steel H, Hetherington S, Pearce G, Spreen W, Lafon S. 2004. Antiviral efficacy of abacavir in antiretroviral therapy-experienced adults harbouring HIV-1 with specific patterns of resistance to nucleoside reverse transcriptase inhibitors. Antivir Ther 9:37–45. 10.1177/135965350400900102. - DOI - PubMed
-
- Kempf DJ, Isaacson JD, King MS, Brun SC, Sylte J, Richards B, Bernstein B, Rode R, Sun E. 2002. Analysis of the virological response with respect to baseline viral phenotype and genotype in protease inhibitor-experienced HIV-1-infected patients receiving lopinavir/ritonavir therapy. Antivir Ther 7:165–174. 10.1177/135965350200700305. - DOI - PubMed

