Genome-wide detection of copy number variation in American mink using whole-genome sequencing
- PMID: 36096727
- PMCID: PMC9468235
- DOI: 10.1186/s12864-022-08874-1
Genome-wide detection of copy number variation in American mink using whole-genome sequencing
Abstract
Background: Copy number variations (CNVs) represent a major source of genetic diversity and contribute to the phenotypic variation of economically important traits in livestock species. In this study, we report the first genome-wide CNV analysis of American mink using whole-genome sequence data from 100 individuals. The analyses were performed by three complementary software programs including CNVpytor, DELLY and Manta.
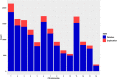
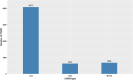
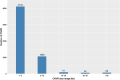
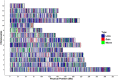
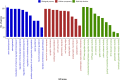
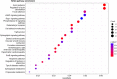
Results: A total of 164,733 CNVs (144,517 deletions and 20,216 duplications) were identified representing 5378 CNV regions (CNVR) after merging overlapping CNVs, covering 47.3 Mb (1.9%) of the mink autosomal genome. Gene Ontology and KEGG pathway enrichment analyses of 1391 genes that overlapped CNVR revealed potential role of CNVs in a wide range of biological, molecular and cellular functions, e.g., pathways related to growth (regulation of actin cytoskeleton, and cAMP signaling pathways), behavior (axon guidance, circadian entrainment, and glutamatergic synapse), lipid metabolism (phospholipid binding, sphingolipid metabolism and regulation of lipolysis in adipocytes), and immune response (Wnt signaling, Fc receptor signaling, and GTPase regulator activity pathways). Furthermore, several CNVR-harbored genes associated with fur characteristics and development (MYO5A, RAB27B, FGF12, SLC7A11, EXOC2), and immune system processes (SWAP70, FYN, ORAI1, TRPM2, and FOXO3).
Conclusions: This study presents the first genome-wide CNV map of American mink. We identified 5378 CNVR in the mink genome and investigated genes that overlapped with CNVR. The results suggest potential links with mink behaviour as well as their possible impact on fur quality and immune response. Overall, the results provide new resources for mink genome analysis, serving as a guideline for future investigations in which genomic structural variations are present.
Keywords: American mink; Copy number variation; Whole-genome sequencing.
© 2022. The Author(s).
Conflict of interest statement
The authors declare they have no competing interests.
Figures
Similar articles
-
Genome-wide association studies for economically important traits in mink using copy number variation.Sci Rep. 2024 Jan 2;14(1):24. doi: 10.1038/s41598-023-50497-3. Sci Rep. 2024. PMID: 38167844 Free PMC article.
-
A global analysis of CNVs in diverse yak populations using whole-genome resequencing.BMC Genomics. 2019 Jan 18;20(1):61. doi: 10.1186/s12864-019-5451-5. BMC Genomics. 2019. PMID: 30658572 Free PMC article.
-
Characterization of runs of homozygosity islands in American mink using whole-genome sequencing data.J Anim Breed Genet. 2024 Sep;141(5):507-520. doi: 10.1111/jbg.12859. Epub 2024 Feb 22. J Anim Breed Genet. 2024. PMID: 38389405
-
A genome-wide analysis of copy number variation in Murciano-Granadina goats.Genet Sel Evol. 2020 Aug 8;52(1):44. doi: 10.1186/s12711-020-00564-4. Genet Sel Evol. 2020. PMID: 32770942 Free PMC article.
-
Advancements in copy number variation screening in herbivorous livestock genomes and their association with phenotypic traits.Front Vet Sci. 2024 Jan 11;10:1334434. doi: 10.3389/fvets.2023.1334434. eCollection 2023. Front Vet Sci. 2024. PMID: 38274664 Free PMC article. Review.
Cited by
-
Genomic Regions Associated with Growth and Reproduction Traits in Pink-Eyed White Mink.Genes (Basel). 2024 Aug 29;15(9):1142. doi: 10.3390/genes15091142. Genes (Basel). 2024. PMID: 39336733 Free PMC article.
-
Whole-genome sequencing of copy number variation analysis in Ethiopian cattle reveals adaptations to diverse environments.BMC Genomics. 2024 Nov 15;25(1):1088. doi: 10.1186/s12864-024-10936-5. BMC Genomics. 2024. PMID: 39548375 Free PMC article.
-
Rosemary extract improves egg quality by altering gut barrier function, intestinal microbiota and oviductal gene expressions in late-phase laying hens.J Anim Sci Biotechnol. 2023 Sep 4;14(1):121. doi: 10.1186/s40104-023-00904-6. J Anim Sci Biotechnol. 2023. PMID: 37667318 Free PMC article.
-
A comprehensive map of copy number variations in dromedary camels based on whole genome sequence data.Sci Rep. 2024 Oct 26;14(1):25573. doi: 10.1038/s41598-024-77773-0. Sci Rep. 2024. PMID: 39462079 Free PMC article.
-
Genome-wide association studies for economically important traits in mink using copy number variation.Sci Rep. 2024 Jan 2;14(1):24. doi: 10.1038/s41598-023-50497-3. Sci Rep. 2024. PMID: 38167844 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

