O-GlcNAc transferase is important for homology-directed repair
- PMID: 36095925
- PMCID: PMC9884008
- DOI: 10.1016/j.dnarep.2022.103394
O-GlcNAc transferase is important for homology-directed repair
Abstract
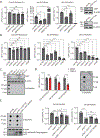
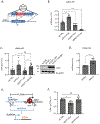
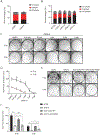
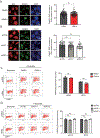
O-Linked β-N-acetylglucosamine glycosylation (O-GlcNAcylation) to serine or threonine residues is a reversible and dynamic post-translational modification. O-GlcNAc transferase (OGT) is the only enzyme for O-GlcNAcylation, and is a potential cancer therapeutic target in combination with clastogenic (i.e., chromosomal breaking) therapeutics. Thus, we sought to examine the influence of O-GlcNAcylation on chromosomal break repair. Using a set of DNA double strand break (DSB) reporter assays, we found that the depletion of OGT, and its inhibition with a small molecule each caused a reduction in repair pathways that involve use of homology: RAD51-dependent homology-directed repair (HDR), and single strand annealing. In contrast, such OGT disruption did not obviously affect chromosomal break end joining, and furthermore caused an increase in homology-directed gene targeting. Such disruption in OGT also caused a reduction in clonogenic survival, as well as modifications to cell cycle profiles, particularly an increase in G1-phase cells. We also examined intermediate steps of HDR, finding no obvious effects on an assay for DSB end resection, nor for RAD51 recruitment into ionizing radiation induced foci (IRIF) in proliferating cells. However, we also found that the influence of OGT on HDR and homology-directed gene targeting were dependent on RAD52, and that OGT is important for RAD52 IRIF in proliferating cells. Thus, we suggest that OGT is important for regulation of HDR that is partially linked to RAD52 function.
Keywords: Double-strand break; Homology-directed repair; O-GlcNAc; OGT; RAD52.
Copyright © 2022 Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that there are no conflicts of interest.
Figures
Similar articles
-
OGT mediated histone H2B S112 GlcNAcylation regulates DNA damage response.J Genet Genomics. 2015 Sep 20;42(9):467-75. doi: 10.1016/j.jgg.2015.07.002. Epub 2015 Jul 23. J Genet Genomics. 2015. PMID: 26408091
-
Elevated O-GlcNAcylation promotes gastric cancer cells proliferation by modulating cell cycle related proteins and ERK 1/2 signaling.Oncotarget. 2016 Sep 20;7(38):61390-61402. doi: 10.18632/oncotarget.11359. Oncotarget. 2016. PMID: 27542217 Free PMC article.
-
O-GlcNAc modification affects the ATM-mediated DNA damage response.Biochim Biophys Acta. 2012 Oct;1820(10):1678-85. doi: 10.1016/j.bbagen.2012.06.013. Epub 2012 Jul 1. Biochim Biophys Acta. 2012. PMID: 22759405
-
Overview of the Assays to Probe O-Linked β-N-Acetylglucosamine Transferase Binding and Activity.Molecules. 2021 Feb 16;26(4):1037. doi: 10.3390/molecules26041037. Molecules. 2021. PMID: 33669256 Free PMC article. Review.
-
'O-GlcNAc Code' Mediated Biological Functions of Downstream Proteins.Molecules. 2018 Aug 6;23(8):1967. doi: 10.3390/molecules23081967. Molecules. 2018. PMID: 30082668 Free PMC article. Review.
Cited by
-
A novel shark single-domain antibody targeting OGT as a tool for detection and intracellular localization.Front Immunol. 2023 Feb 10;14:1062656. doi: 10.3389/fimmu.2023.1062656. eCollection 2023. Front Immunol. 2023. PMID: 36855630 Free PMC article.
-
Extracellular arginine availability modulates eIF2α O-GlcNAcylation and heme oxygenase 1 translation for cellular homeostasis.J Biomed Sci. 2023 May 22;30(1):32. doi: 10.1186/s12929-023-00924-4. J Biomed Sci. 2023. PMID: 37217939 Free PMC article.
-
Drug Discovery Targeting Post-Translational Modifications in Response to DNA Damages Induced by Space Radiation.Int J Mol Sci. 2023 Apr 21;24(8):7656. doi: 10.3390/ijms24087656. Int J Mol Sci. 2023. PMID: 37108815 Free PMC article. Review.
-
RAD18 O-GlcNAcylation promotes translesion DNA synthesis and homologous recombination repair.Cell Death Dis. 2024 May 8;15(5):321. doi: 10.1038/s41419-024-06700-y. Cell Death Dis. 2024. PMID: 38719812 Free PMC article.
References
-
- Dong DLY, Hart GW, Purification and Characterization of an O-Glcnac Selective N-Acetyl-Beta-D-Glucosaminidase from Rat Spleen Cytosol, J Biol Chem, 269 (1994) 19321–19330. - PubMed
-
- Hanover JA, Yu S, Lubas WB, Shin SH, Ragano-Caracciola M, Kochran J, Love DC, Mitochondrial and nucleocytoplasmic isoforms of O-linked GlcNAc transferase encoded by a single mammalian gene, Arch Biochem Biophys, 409 (2003) 287–297. - PubMed
-
- Haltiwanger RS, Holt GD, Hart GW, Enzymatic Addition of O-Glcnac to Nuclear and Cytoplasmic Proteins - Identification of a Uridine Diphospho-N-Acetylglucosamine-Peptide Beta-N-Acetylglucosaminyltransferase, J Biol Chem, 265 (1990) 2563–2568. - PubMed
-
- Hart GW, Housley MP, Slawson C, Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins, Nature, 446 (2007) 1017–1022. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

