Molecular basis of epigenetic regulation in cancer diagnosis and treatment
- PMID: 36092905
- PMCID: PMC9449878
- DOI: 10.3389/fgene.2022.885635
Molecular basis of epigenetic regulation in cancer diagnosis and treatment
Abstract
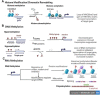
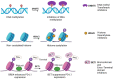
The global cancer cases and mortality rates are increasing and demand efficient biomarkers for accurate screening, detection, diagnosis, and prognosis. Recent studies have demonstrated that variations in epigenetic mechanisms like aberrant promoter methylation, altered histone modification and mutations in ATP-dependent chromatin remodelling complexes play an important role in the development of carcinogenic events. However, the influence of other epigenetic alterations in various cancers was confirmed with evolving research and the emergence of high throughput technologies. Therefore, alterations in epigenetic marks may have clinical utility as potential biomarkers for early cancer detection and diagnosis. In this review, an outline of the key epigenetic mechanism(s), and their deregulation in cancer etiology have been discussed to decipher the future prospects in cancer therapeutics including precision medicine. Also, this review attempts to highlight the gaps in epigenetic drug development with emphasis on integrative analysis of epigenetic biomarkers to establish minimally non-invasive biomarkers with clinical applications.
Keywords: DNA methylation; cancer; cancer therapies; chromatin remodelling; epigenetics; histone modification.
Copyright © 2022 Tulsyan, Aftab, Sisodiya, Khan, Chikara, Tanwar and Hussain.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a no conflict of interest.
Figures



Similar articles
-
Epigenetics and cancer, 2nd IARC meeting, Lyon, France, 6 and 7 December 2007.Mol Oncol. 2008 Jun;2(1):33-40. doi: 10.1016/j.molonc.2008.03.005. Epub 2008 Mar 27. Mol Oncol. 2008. PMID: 19383327 Free PMC article.
-
Role of epigenetics in carcinogenesis: Recent advancements in anticancer therapy.Semin Cancer Biol. 2022 Aug;83:441-451. doi: 10.1016/j.semcancer.2021.06.023. Epub 2021 Jun 26. Semin Cancer Biol. 2022. PMID: 34182144 Review.
-
Early Epigenetic Markers for Precision Medicine.Methods Mol Biol. 2018;1856:3-17. doi: 10.1007/978-1-4939-8751-1_1. Methods Mol Biol. 2018. PMID: 30178243 Review.
-
Progresses in epigenetic studies of asthma from the perspective of high-throughput analysis technologies: a narrative review.Ann Transl Med. 2022 Apr;10(8):493. doi: 10.21037/atm-22-929. Ann Transl Med. 2022. PMID: 35571411 Free PMC article. Review.
-
Pharmaco-epigenomics: On the Road of Translation Medicine.Adv Exp Med Biol. 2019;1168:31-42. doi: 10.1007/978-3-030-24100-1_3. Adv Exp Med Biol. 2019. PMID: 31713163 Review.
Cited by
-
Pancreatic Neuroendocrine Tumors: Signaling Pathways and Epigenetic Regulation.Int J Mol Sci. 2024 Jan 22;25(2):1331. doi: 10.3390/ijms25021331. Int J Mol Sci. 2024. PMID: 38279330 Free PMC article. Review.
-
Exploring the promise of regulator of G Protein Signaling 20: insights into potential mechanisms and prospects across solid cancers and hematological malignancies.Cancer Cell Int. 2024 Sep 3;24(1):305. doi: 10.1186/s12935-024-03487-y. Cancer Cell Int. 2024. PMID: 39227952 Free PMC article. Review.
-
Insights into the Mechanism of Curaxin CBL0137 Epigenetic Activity: The Induction of DNA Demethylation and the Suppression of BET Family Proteins.Int J Mol Sci. 2023 Aug 17;24(16):12874. doi: 10.3390/ijms241612874. Int J Mol Sci. 2023. PMID: 37629054 Free PMC article.
-
Epigenetics: Mechanisms, potential roles, and therapeutic strategies in cancer progression.Genes Dis. 2023 Jul 6;11(5):101020. doi: 10.1016/j.gendis.2023.04.040. eCollection 2024 Sep. Genes Dis. 2023. PMID: 38988323 Free PMC article. Review.
-
Epigenetic modulation of ferroptosis in cancer: Identifying epigenetic targets for novel anticancer therapy.Cell Oncol (Dordr). 2023 Dec;46(6):1605-1623. doi: 10.1007/s13402-023-00840-7. Epub 2023 Jul 12. Cell Oncol (Dordr). 2023. PMID: 37438601 Review.
References
Publication types
LinkOut - more resources
Full Text Sources

