Peroxisomal very long-chain fatty acid transport is targeted by herpesviruses and the antiviral host response
- PMID: 36085307
- PMCID: PMC9462615
- DOI: 10.1038/s42003-022-03867-y
Peroxisomal very long-chain fatty acid transport is targeted by herpesviruses and the antiviral host response
Abstract
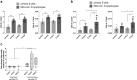
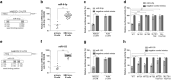
Very long-chain fatty acids (VLCFA) are critical for human cytomegalovirus replication and accumulate upon infection. Here, we used Epstein-Barr virus (EBV) infection of human B cells to elucidate how herpesviruses target VLCFA metabolism. Gene expression profiling revealed that, despite a general induction of peroxisome-related genes, EBV early infection decreased expression of the peroxisomal VLCFA transporters ABCD1 and ABCD2, thus impairing VLCFA degradation. The mechanism underlying ABCD1 and ABCD2 repression involved RNA interference by the EBV-induced microRNAs miR-9-5p and miR-155, respectively, causing significantly increased VLCFA levels. Treatment with 25-hydroxycholesterol, an antiviral innate immune modulator produced by macrophages, restored ABCD1 expression and reduced VLCFA accumulation in EBV-infected B-lymphocytes, and, upon lytic reactivation, reduced virus production in control but not ABCD1-deficient cells. Finally, also other herpesviruses and coronaviruses target ABCD1 expression. Because viral infection might trigger neuroinflammation in X-linked adrenoleukodystrophy (X-ALD, inherited ABCD1 deficiency), we explored a possible link between EBV infection and cerebral X-ALD. However, neither immunohistochemistry of post-mortem brains nor analysis of EBV seropositivity in 35 X-ALD children supported involvement of EBV in the onset of neuroinflammation. Collectively, our findings indicate a previously unrecognized, pivotal role of ABCD1 in viral infection and host defence, prompting consideration of other viral triggers in cerebral X-ALD.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Saturated very long-chain fatty acids regulate macrophage plasticity and invasiveness.J Neuroinflammation. 2022 Dec 17;19(1):305. doi: 10.1186/s12974-022-02664-y. J Neuroinflammation. 2022. PMID: 36528616 Free PMC article.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Epstein-Barr Virus Envelope Glycoprotein gp110 Inhibits IKKi-Mediated Activation of NF-κB and Promotes the Degradation of β-Catenin.Microbiol Spectr. 2023 Jun 15;11(3):e0032623. doi: 10.1128/spectrum.00326-23. Epub 2023 Apr 6. Microbiol Spectr. 2023. PMID: 37022262 Free PMC article.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article. Review.
Cited by
-
Efficacy of HDAC Inhibitors in Driving Peroxisomal β-Oxidation and Immune Responses in Human Macrophages: Implications for Neuroinflammatory Disorders.Biomolecules. 2023 Nov 23;13(12):1696. doi: 10.3390/biom13121696. Biomolecules. 2023. PMID: 38136568 Free PMC article.
-
Peroxisomal Localization of a Truncated HMG-CoA Reductase under Low Cholesterol Conditions.Biomolecules. 2024 Feb 19;14(2):244. doi: 10.3390/biom14020244. Biomolecules. 2024. PMID: 38397481 Free PMC article.
-
Neuroproteomic Analysis after SARS-CoV-2 Infection Reveals Overrepresented Neurodegeneration Pathways and Disrupted Metabolic Pathways.Biomolecules. 2023 Oct 30;13(11):1597. doi: 10.3390/biom13111597. Biomolecules. 2023. PMID: 38002279 Free PMC article.
-
Herpesviral interplay with peroxisome: An underexplored viral niche.Genes Dis. 2023 Mar 24;10(4):1133-1135. doi: 10.1016/j.gendis.2023.01.033. eCollection 2023 Jul. Genes Dis. 2023. PMID: 37397512 Free PMC article.
-
Saturated very long-chain fatty acids regulate macrophage plasticity and invasiveness.J Neuroinflammation. 2022 Dec 17;19(1):305. doi: 10.1186/s12974-022-02664-y. J Neuroinflammation. 2022. PMID: 36528616 Free PMC article.
References
-
- Bjornevik, K. et al. Longitudinal analysis reveals high prevalence of Epstein-Barr virus associated with multiple sclerosis. Science10.1126/science.abj8222 (2022). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

