Structural insights into human CCAN complex assembled onto DNA
- PMID: 36085283
- PMCID: PMC9463443
- DOI: 10.1038/s41421-022-00439-6
Structural insights into human CCAN complex assembled onto DNA
Abstract
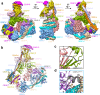
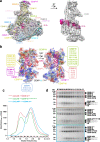
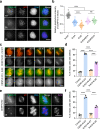
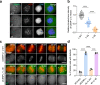
In mitosis, accurate chromosome segregation depends on kinetochores that connect centromeric chromatin to spindle microtubules. The centromeres of budding yeast, which are relatively simple, are connected to individual microtubules via a kinetochore constitutive centromere associated network (CCAN). However, the complex centromeres of human chromosomes comprise millions of DNA base pairs and attach to multiple microtubules. Here, by use of cryo-electron microscopy and functional analyses, we reveal the molecular basis of how human CCAN interacts with duplex DNA and facilitates accurate chromosome segregation. The overall structure relates to the cooperative interactions and interdependency of the constituent sub-complexes of the CCAN. The duplex DNA is topologically entrapped by human CCAN. Further, CENP-N does not bind to the RG-loop of CENP-A but to DNA in the CCAN complex. The DNA binding activity is essential for CENP-LN localization to centromere and chromosome segregation during mitosis. Thus, these analyses provide new insights into mechanisms of action underlying kinetochore assembly and function in mitosis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome.Nature. 2019 Oct;574(7777):278-282. doi: 10.1038/s41586-019-1609-1. Epub 2019 Oct 2. Nature. 2019. PMID: 31578520 Free PMC article.
-
Conserved and divergent mechanisms of inner kinetochore assembly onto centromeric chromatin.Curr Opin Struct Biol. 2023 Aug;81:102638. doi: 10.1016/j.sbi.2023.102638. Epub 2023 Jun 20. Curr Opin Struct Biol. 2023. PMID: 37343495 Review.
-
Centromere/kinetochore is assembled through CENP-C oligomerization.Mol Cell. 2023 Jul 6;83(13):2188-2205.e13. doi: 10.1016/j.molcel.2023.05.023. Epub 2023 Jun 8. Mol Cell. 2023. PMID: 37295434
-
Structure of the human inner kinetochore CCAN complex and its significance for human centromere organization.Mol Cell. 2022 Jun 2;82(11):2113-2131.e8. doi: 10.1016/j.molcel.2022.04.027. Epub 2022 May 6. Mol Cell. 2022. PMID: 35525244 Free PMC article.
-
An updated view of the kinetochore architecture.Trends Genet. 2023 Dec;39(12):941-953. doi: 10.1016/j.tig.2023.09.003. Epub 2023 Sep 27. Trends Genet. 2023. PMID: 37775394 Review.
Cited by
-
Dynamic phosphorylation of CENP-N by CDK1 guides accurate chromosome segregation in mitosis.J Mol Cell Biol. 2023 Nov 27;15(6):mjad041. doi: 10.1093/jmcb/mjad041. J Mol Cell Biol. 2023. PMID: 37365681 Free PMC article.
-
PLK1 phosphorylation of ZW10 guides accurate chromosome segregation in mitosis.J Mol Cell Biol. 2024 Jul 29;16(2):mjae008. doi: 10.1093/jmcb/mjae008. J Mol Cell Biol. 2024. PMID: 38402459 Free PMC article.
-
A non-canonical role of the inner kinetochore in regulating sister-chromatid cohesion at centromeres.EMBO J. 2024 Jun;43(12):2424-2452. doi: 10.1038/s44318-024-00104-6. Epub 2024 May 7. EMBO J. 2024. PMID: 38714893 Free PMC article.
-
Sgo1 interacts with CENP-A to guide accurate chromosome segregation in mitosis.J Mol Cell Biol. 2024 Apr 4;15(10):mjad061. doi: 10.1093/jmcb/mjad061. J Mol Cell Biol. 2024. PMID: 37777834 Free PMC article.
-
CENPA functions as a transcriptional regulator to promote hepatocellular carcinoma progression via cooperating with YY1.Int J Biol Sci. 2023 Oct 16;19(16):5218-5232. doi: 10.7150/ijbs.85656. eCollection 2023. Int J Biol Sci. 2023. PMID: 37928273 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

