Quantum Biochemistry and MM-PBSA Description of the ZIKV NS2B-NS3 Protease: Insights into the Binding Interactions beyond the Catalytic Triad Pocket
- PMID: 36077486
- PMCID: PMC9456192
- DOI: 10.3390/ijms231710088
Quantum Biochemistry and MM-PBSA Description of the ZIKV NS2B-NS3 Protease: Insights into the Binding Interactions beyond the Catalytic Triad Pocket
Abstract
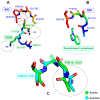
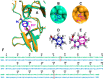
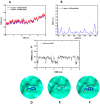
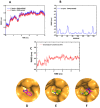
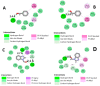
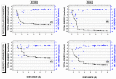
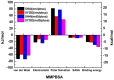
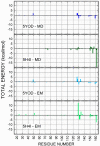
The Zika virus protease NS2B-NS3 has a binding site formed with the participation of a H51-D75-S135 triad presenting two forms, active and inactive. Studies suggest that the inactive conformation is a good target for the design of inhibitors. In this paper, we evaluated the co-crystallized structures of the protease with the inhibitors benzoic acid (5YOD) and benzimidazole-1-ylmethanol (5H4I). We applied a protocol consisting of two steps: first, classical molecular mechanics energy minimization followed by classical molecular dynamics were performed, obtaining stabilized molecular geometries; second, the optimized/relaxed geometries were used in quantum biochemistry and molecular mechanics/Poisson-Boltzmann surface area (MM-PBSA) calculations to estimate the ligand interactions with each amino acid residue of the binding pocket. We show that the quantum-level results identified essential residues for the stabilization of the 5YOD and 5H4I complexes after classical energy minimization, matching previously published experimental data. The same success, however, was not observed for the MM-PBSA simulations. The application of quantum biochemistry methods seems to be more promising for the design of novel inhibitors acting on NS2B-NS3.
Keywords: MM-PBSA; NS2B-NS3; Zika virus; oxyanion orifice; quantum biochemistry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
MD simulations reveal alternate conformations of the oxyanion hole in the Zika virus NS2B/NS3 protease.Proteins. 2020 Feb;88(2):345-354. doi: 10.1002/prot.25809. Epub 2019 Sep 9. Proteins. 2020. PMID: 31461176
-
pH and non-covalent ligand binding modulate Zika virus NS2B/NS3 protease binding site residues: Discoveries from MD and constant pH MD simulations.J Biomol Struct Dyn. 2022;40(20):10359-10372. doi: 10.1080/07391102.2021.1943528. Epub 2021 Jun 27. J Biomol Struct Dyn. 2022. PMID: 34180376
-
Allosteric Inhibitors of Zika Virus NS2B-NS3 Protease Targeting Protease in "Super-Open" Conformation.Viruses. 2023 Apr 30;15(5):1106. doi: 10.3390/v15051106. Viruses. 2023. PMID: 37243192 Free PMC article.
-
The Structure of the Zika Virus Protease, NS2B/NS3pro.Adv Exp Med Biol. 2018;1062:131-145. doi: 10.1007/978-981-10-8727-1_10. Adv Exp Med Biol. 2018. PMID: 29845530 Review.
-
Exploiting the unique features of Zika and Dengue proteases for inhibitor design.Biochimie. 2019 Nov;166:132-141. doi: 10.1016/j.biochi.2019.05.004. Epub 2019 May 9. Biochimie. 2019. PMID: 31077760 Review.
Cited by
-
ZIKV Inhibitors Based on Pyrazolo[3,4-d]pyridazine-7-one Core: Rational Design, In Vitro Evaluation, and Theoretical Studies.ACS Omega. 2023 Dec 14;8(51):48994-49008. doi: 10.1021/acsomega.3c06612. eCollection 2023 Dec 26. ACS Omega. 2023. PMID: 38162759 Free PMC article.
-
Recent Advances in Biochemistry and Molecular Biology of Infectious Diseases.Int J Mol Sci. 2023 May 18;24(10):8958. doi: 10.3390/ijms24108958. Int J Mol Sci. 2023. PMID: 37240305 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

