A Direct Infusion Probe for Rapid Metabolomics of Low-Volume Samples
- PMID: 36070505
- PMCID: PMC9494293
- DOI: 10.1021/acs.analchem.2c02918
A Direct Infusion Probe for Rapid Metabolomics of Low-Volume Samples
Abstract
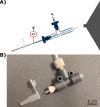
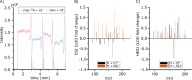
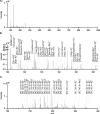
Targeted and nontargeted metabolomics has the potential to evaluate and detect global metabolite changes in biological systems. Direct infusion mass spectrometric analysis enables detection of all ionizable small molecules, thus simultaneously providing information on both metabolites and lipids in chemically complex samples. However, to unravel the heterogeneity of the metabolic status of cells in culture and tissue a low number of cells per sample should be analyzed with high sensitivity, which requires low sample volumes. Here, we present the design and characterization of the direct infusion probe, DIP. The DIP is simple to build and position directly in front of a mass spectrometer for rapid metabolomics of chemically complex biological samples using pneumatically assisted electrospray ionization at 1 μL/min flow rate. The resulting data is acquired in a square wave profile with minimal carryover between samples that enhances throughput and enables several minutes of uniform MS signal from 5 μL sample volumes. The DIP was applied to study the intracellular metabolism of insulin secreting INS-1 cells and the results show that exposure to 20 mM glucose for 15 min significantly alters the abundance of several small metabolites, amino acids, and lipids.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Global and Spatial Metabolomics of Individual Cells Using a Tapered Pneumatically Assisted nano-DESI Probe.J Am Soc Mass Spectrom. 2023 Nov 1;34(11):2518-2524. doi: 10.1021/jasms.3c00239. Epub 2023 Oct 13. J Am Soc Mass Spectrom. 2023. PMID: 37830184 Free PMC article.
-
Translational Metabolomics of Head Injury: Exploring Dysfunctional Cerebral Metabolism with Ex Vivo NMR Spectroscopy-Based Metabolite Quantification.In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. In: Kobeissy FH, editor. Brain Neurotrauma: Molecular, Neuropsychological, and Rehabilitation Aspects. Boca Raton (FL): CRC Press/Taylor & Francis; 2015. Chapter 25. PMID: 26269925 Free Books & Documents. Review.
-
Comparison of liquid chromatography-mass spectrometry and direct infusion microchip electrospray ionization mass spectrometry in global metabolomics of cell samples.Eur J Pharm Sci. 2019 Oct 1;138:104991. doi: 10.1016/j.ejps.2019.104991. Epub 2019 Aug 9. Eur J Pharm Sci. 2019. PMID: 31404622
-
A complete workflow for high-resolution spectral-stitching nanoelectrospray direct-infusion mass-spectrometry-based metabolomics and lipidomics.Nat Protoc. 2016 Feb;12(2):310–328. doi: 10.1038/nprot.2016.156. Epub 2017 Jan 12. Nat Protoc. 2016. PMID: 28079878
-
Recent Advances in Mass Spectrometry-Based Spatially Resolved Molecular Imaging of Drug Disposition and Metabolomics.Drug Metab Dispos. 2023 Oct;51(10):1273-1283. doi: 10.1124/dmd.122.001069. Epub 2023 Jun 9. Drug Metab Dispos. 2023. PMID: 37295949 Review.
Cited by
-
Global and Spatial Metabolomics of Individual Cells Using a Tapered Pneumatically Assisted nano-DESI Probe.J Am Soc Mass Spectrom. 2023 Nov 1;34(11):2518-2524. doi: 10.1021/jasms.3c00239. Epub 2023 Oct 13. J Am Soc Mass Spectrom. 2023. PMID: 37830184 Free PMC article.
-
Recent Analytical Methodologies in Lipid Analysis.Int J Mol Sci. 2024 Feb 13;25(4):2249. doi: 10.3390/ijms25042249. Int J Mol Sci. 2024. PMID: 38396926 Free PMC article. Review.
-
Screening autism-associated environmental factors in differentiating human neural progenitors with fractional factorial design-based transcriptomics.Sci Rep. 2023 Jun 29;13(1):10519. doi: 10.1038/s41598-023-37488-0. Sci Rep. 2023. PMID: 37386098 Free PMC article.
-
Metabolomics-driven approaches for identifying therapeutic targets in drug discovery.MedComm (2020). 2024 Nov 11;5(11):e792. doi: 10.1002/mco2.792. eCollection 2024 Nov. MedComm (2020). 2024. PMID: 39534557 Free PMC article. Review.
References
-
- Dutta M.; Singh B.; Joshi M.; Das D.; Subramani E.; Maan M.; Jana S. K.; Sharma U.; Das S.; Dasgupta S.; Ray C. D.; Chakravarty B.; Chaudhury K. Metabolomics Reveals Perturbations in Endometrium and Serum of Minimal and Mild Endometriosis. Sci. Rep. 2018, 8 (1), 1–9. 10.1038/s41598-018-23954-7. - DOI - PMC - PubMed
-
- Coene K. L. M.; Kluijtmans L. A. J.; van der Heeft E.; Engelke U. F. H.; de Boer S.; Hoegen B.; Kwast H. J. T.; van de Vorst M.; Huigen M. C. D. G.; Keularts I. M. L. W.; Schreuder M. F.; van Karnebeek C. D. M.; Wortmann S. B.; de Vries M. C.; Janssen M. C. H.; Gilissen C.; Engel J.; Wevers R. A. Next-Generation Metabolic Screening: Targeted and Untargeted Metabolomics for the Diagnosis of Inborn Errors of Metabolism in Individual Patients. J. Inherited Metab. Dis. 2018, 41 (3), 337–353. 10.1007/s10545-017-0131-6. - DOI - PMC - PubMed
-
- You X.; Jiang W.; Lu W.; Zhang H.; Yu T.; Tian J.; Wen S.; Garcia-Manero G.; Huang P.; Hu Y. Metabolic Reprogramming and Redox Adaptation in Sorafenib-Resistant Leukemia Cells: Detected by Untargeted Metabolomics and Stable Isotope Tracing Analysis. Cancer Commun. 2019, 39 (1), 17.10.1186/s40880-019-0362-z. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

