Exploring the eukaryotic Yip and REEP/Yop superfamily of membrane-shaping adapter proteins (MSAPs): A cacophony or harmony of structure and function?
- PMID: 36060263
- PMCID: PMC9437294
- DOI: 10.3389/fmolb.2022.912848
Exploring the eukaryotic Yip and REEP/Yop superfamily of membrane-shaping adapter proteins (MSAPs): A cacophony or harmony of structure and function?
Abstract
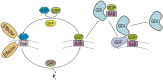
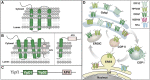
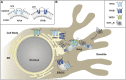
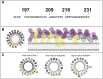
Polytopic cargo proteins are synthesized and exported along the secretory pathway from the endoplasmic reticulum (ER), through the Golgi apparatus, with eventual insertion into the plasma membrane (PM). While searching for proteins that could enhance cell surface expression of olfactory receptors, a new family of proteins termed "receptor expression-enhancing proteins" or REEPs were identified. These membrane-shaping hairpin proteins serve as adapters, interacting with intracellular transport machinery, to regulate cargo protein trafficking. However, REEPs belong to a larger family of proteins, the Yip (Ypt-interacting protein) family, conserved in yeast and higher eukaryotes. To date, eighteen mammalian Yip family members, divided into four subfamilies (Yipf, REEP, Yif, and PRAF), have been identified. Yeast research has revealed many intriguing aspects of yeast Yip function, functions that have not completely been explored with mammalian Yip family members. This review and analysis will clarify the different Yip family nomenclature that have encumbered prior comparisons between yeast, plants, and eukaryotic family members, to provide a more complete understanding of their interacting proteins, membrane topology, organelle localization, and role as regulators of cargo trafficking and localization. In addition, the biological role of membrane shaping and sensing hairpin and amphipathic helical domains of various Yip proteins and their potential cellular functions will be described. Lastly, this review will discuss the concept of Yip proteins as members of a larger superfamily of membrane-shaping adapter proteins (MSAPs), proteins that both shape membranes via membrane-sensing and hairpin insertion, and well as act as adapters for protein-protein interactions. MSAPs are defined by their localization to specific membranes, ability to alter membrane structure, interactions with other proteins via specific domains, and specific interactions/effects on cargo proteins.
Keywords: PRAF; REEP; YIPF; Yif; endoplasmic reticulum; golgi; hereditary spastic paraplegia; membrane-shaping adapter protein.
Copyright © 2022 Angelotti.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
REEPs are membrane shaping adapter proteins that modulate specific g protein-coupled receptor trafficking by affecting ER cargo capacity.PLoS One. 2013 Oct 2;8(10):e76366. doi: 10.1371/journal.pone.0076366. eCollection 2013. PLoS One. 2013. PMID: 24098485 Free PMC article.
-
Arl6IP1 has the ability to shape the mammalian ER membrane in a reticulon-like fashion.Biochem J. 2014 Feb 15;458(1):69-79. doi: 10.1042/BJ20131186. Biochem J. 2014. PMID: 24262037
-
The REEP family of proteins: Molecular targets and role in pathophysiology.Pharmacol Res. 2022 Nov;185:106477. doi: 10.1016/j.phrs.2022.106477. Epub 2022 Sep 30. Pharmacol Res. 2022. PMID: 36191880 Review.
-
TRP Channel Trafficking.In: Liedtke WB, Heller S, editors. TRP Ion Channel Function in Sensory Transduction and Cellular Signaling Cascades. Boca Raton (FL): CRC Press/Taylor & Francis; 2007. Chapter 23. In: Liedtke WB, Heller S, editors. TRP Ion Channel Function in Sensory Transduction and Cellular Signaling Cascades. Boca Raton (FL): CRC Press/Taylor & Francis; 2007. Chapter 23. PMID: 21204515 Free Books & Documents. Review.
-
REEP1 and REEP2 proteins are preferentially expressed in neuronal and neuronal-like exocytotic tissues.Brain Res. 2014 Jan 30;1545:12-22. doi: 10.1016/j.brainres.2013.12.008. Epub 2013 Dec 16. Brain Res. 2014. PMID: 24355597 Free PMC article.
Cited by
-
Molecular Responses of Anti-VEGF Therapy in Neovascular Age-Related Macular Degeneration: Integrative Insights From Multi-Omics and Clinical Imaging.Invest Ophthalmol Vis Sci. 2024 Aug 1;65(10):24. doi: 10.1167/iovs.65.10.24. Invest Ophthalmol Vis Sci. 2024. PMID: 39140961 Free PMC article.
-
YIPF2 regulates genome integrity.Cell Biosci. 2024 Sep 5;14(1):114. doi: 10.1186/s13578-024-01300-x. Cell Biosci. 2024. PMID: 39238039 Free PMC article.
-
AtHVA22a, a plant-specific homologue of Reep/DP1/Yop1 family proteins is involved in turnip mosaic virus propagation.Mol Plant Pathol. 2024 May;25(5):e13466. doi: 10.1111/mpp.13466. Mol Plant Pathol. 2024. PMID: 38767756 Free PMC article.
References
-
- Al Awabdh S., Miserey-Lenkei S., Bouceba T., Masson J., Kano F., Marinach-Patrice C., et al. (2012). A new vesicular scaffolding complex mediates the G-protein-coupled 5-ht1a receptor targeting to neuronal dendrites. J. Neurosci. 32 (41), 14227–14241. 10.1523/jneurosci.6329-11.2012 - DOI - PMC - PubMed
-
- AlMuhaizea M., AlMass R., AlHargan A., AlBader A., Medico Salsench E., Howaidi J., et al. (2020). Truncating mutations in YIF1B cause a progressive encephalopathy with various degrees of mixed movement disorder, microcephaly, and epilepsy. Acta Neuropathol. 139 (4), 791–794. 10.1007/s00401-020-02128-8 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

