Integrated analysis of the expression profiles of the lncRNA-miRNA-mRNA ceRNA network in granulosa and cumulus cells from yak ovaries
- PMID: 36057545
- PMCID: PMC9441039
- DOI: 10.1186/s12864-022-08848-3
Integrated analysis of the expression profiles of the lncRNA-miRNA-mRNA ceRNA network in granulosa and cumulus cells from yak ovaries
Abstract
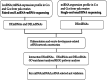
Background: Growing oocytes acquire the ability to mature through two-way communication between gametes and surrounding somatic cumulus cells (CCs). Granulosa cells (GCs) support oocyte growth, regulate meiosis progression, and modulate global oocyte transcription activity. However, the proliferation and differentiation of the yak ovary in GCs and CCs remain unclear. To characterize the important roles of long non-coding RNA, (lncRNA), microRNA (miRNA), and messenger RNA (mRNA), whole-transcriptome analysis was performed. Real-time quantitative fluorescence PCR was performed to verify the selected RNA sequences.
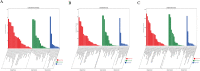
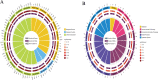
Results: Important gene ontology terms and Kyoto Encyclopedia of Genes and Genomes pathways related to differentiation and oocyte development were identified for the target genes of differentially expressed lncRNAs, miRNAs, and mRNAs. In total,6223 mRNAs (2197 upregulated, 4026 downregulated), 643 lncRNAs (204 upregulated, 479 downregulated), and 559 miRNAs (311 upregulated, 248 downregulated) were significantly altered between the two groups. Target genes involved in cell adhesion, cell differentiation, regulation of developmental processes, cell proliferation, embryo development, signal transduction, apoptosis, and aromatic compound biosynthetic processes were significantly enriched. These RNAs were involved in ECM-receptor interaction, MAPK signaling, Hippo signaling, PI3K-Akt signaling, cell cycle, cell adhesion, leukocyte trans-endothelial migration, and actin cytoskeleton regulation.
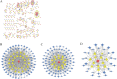
Conclusions: A comprehensive analysis of the co-expression network of competing endogenous RNAs (ceRNAs) will facilitate the understanding of the process of granulosa cell proliferation and differentiation and offer a theoretical basis for the development of oocytes.
Keywords: Cumulus cells; Granulosa cells; LncRNA; ceRNA network; miRNA.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Comprehensive analysis of lncRNA-miRNA-mRNA networks during osteogenic differentiation of bone marrow mesenchymal stem cells.BMC Genomics. 2022 Jun 7;23(1):425. doi: 10.1186/s12864-022-08646-x. BMC Genomics. 2022. PMID: 35672672 Free PMC article.
-
Down-regulation of long non-coding RNAs in reproductive aging and analysis of the lncRNA-miRNA-mRNA networks in human cumulus cells.J Assist Reprod Genet. 2022 Apr;39(4):919-931. doi: 10.1007/s10815-022-02446-8. Epub 2022 Mar 5. J Assist Reprod Genet. 2022. PMID: 35247118 Free PMC article.
-
The long non-coding RNA, miRNA and mRNA landscapes of cementoblasts during cementogenesis.Orthod Craniofac Res. 2023 Nov;26(4):667-678. doi: 10.1111/ocr.12668. Epub 2023 May 2. Orthod Craniofac Res. 2023. PMID: 37129094
-
Reconstruction and Analysis of the Differentially Expressed IncRNA-miRNA-mRNA Network Based on Competitive Endogenous RNA in Hepatocellular Carcinoma.Crit Rev Eukaryot Gene Expr. 2019;29(6):539-549. doi: 10.1615/CritRevEukaryotGeneExpr.2019028740. Crit Rev Eukaryot Gene Expr. 2019. PMID: 32422009 Review.
-
Long noncoding RNA SNHG7-miRNA-mRNA axes crosstalk with oncogenic signaling pathways in human cancers.Chem Biol Drug Des. 2023 May;101(5):1151-1161. doi: 10.1111/cbdd.14118. Epub 2022 Aug 22. Chem Biol Drug Des. 2023. PMID: 35993390 Review.
Cited by
-
Transcriptomics reveals age-related changes in ion transport-related factors in yak lungs.Front Vet Sci. 2024 May 8;11:1374794. doi: 10.3389/fvets.2024.1374794. eCollection 2024. Front Vet Sci. 2024. PMID: 38779034 Free PMC article.
-
Transcriptomics and metabolomics of blood, urine and ovarian follicular fluid of yak at induced estrus stage.BMC Genomics. 2024 Feb 21;25(1):201. doi: 10.1186/s12864-024-10079-7. BMC Genomics. 2024. PMID: 38383305 Free PMC article.
-
Transcriptome Analysis Provides Insights into Copulation, Fertilization, and Gestation in Sebastes schlegelii.Genes (Basel). 2022 Oct 7;13(10):1812. doi: 10.3390/genes13101812. Genes (Basel). 2022. PMID: 36292697 Free PMC article.
-
Exosomes Derived from Yak Follicular Fluid Increase 2-Hydroxyestradiol Secretion by Activating Autophagy in Cumulus Cells.Animals (Basel). 2022 Nov 16;12(22):3174. doi: 10.3390/ani12223174. Animals (Basel). 2022. PMID: 36428401 Free PMC article.
References
-
- Macklon NS, Fauser B. Aspects of ovarian follicle development throughout life. Horm Res. 2000;52(4):161–170. - PubMed
-
- Vanderhyden T. Eppig: mouse oocytes promote proliferation of granulosa cells from preantral and antral follicles in vitro. Biol Reprod. 1992;46(6):1196–204. - PubMed
-
- Kempisty B, Ziółkowska A, Ciesiółka S, Piotrowska H, Antosik P, Bukowska D, Nowicki M, Brüssow KP, Zabel M. Study on connexin gene and protein expression and cellular distribution in relation to real-time proliferation of porcine granulosa cells. J Biol Regul Homeost Agents. 2014;28(4):625–635. - PubMed
MeSH terms
Substances
Grants and funding
- 31972760/National Natural Science Foundation of China
- 31972760/National Natural Science Foundation of China
- 31972760/National Natural Science Foundation of China
- 31972760/National Natural Science Foundation of China
- 31972760/National Natural Science Foundation of China
- 31972760/National Natural Science Foundation of China
- 31972760/National Natural Science Foundation of China
- 31972760/National Natural Science Foundation of China
- 31972760/National Natural Science Foundation of China
- 31972760/National Natural Science Foundation of China
- 31972760/National Natural Science Foundation of China
- 31972760/National Natural Science Foundation of China
- 20JR10RA561/Prominent Youth Foundation of Gansu Province
- 20JR10RA561/Prominent Youth Foundation of Gansu Province
- 20JR10RA561/Prominent Youth Foundation of Gansu Province
- 20JR10RA561/Prominent Youth Foundation of Gansu Province
- 20JR10RA561/Prominent Youth Foundation of Gansu Province
- 20JR10RA561/Prominent Youth Foundation of Gansu Province
- 20JR10RA561/Prominent Youth Foundation of Gansu Province
- 20JR10RA561/Prominent Youth Foundation of Gansu Province
- 20JR10RA561/Prominent Youth Foundation of Gansu Province
- 20JR10RA561/Prominent Youth Foundation of Gansu Province
- 20JR10RA561/Prominent Youth Foundation of Gansu Province
- 20JR10RA561/Prominent Youth Foundation of Gansu Province
- 21ZD10NA001/Major Science and Technology Project of Gansu Province
- 21ZD10NA001/Major Science and Technology Project of Gansu Province
- 21ZD10NA001/Major Science and Technology Project of Gansu Province
- 21ZD10NA001/Major Science and Technology Project of Gansu Province
- 21ZD10NA001/Major Science and Technology Project of Gansu Province
- 21ZD10NA001/Major Science and Technology Project of Gansu Province
- 21ZD10NA001/Major Science and Technology Project of Gansu Province
- 21ZD10NA001/Major Science and Technology Project of Gansu Province
- 21ZD10NA001/Major Science and Technology Project of Gansu Province
- 21ZD10NA001/Major Science and Technology Project of Gansu Province
- 21ZD10NA001/Major Science and Technology Project of Gansu Province
- 21ZD10NA001/Major Science and Technology Project of Gansu Province
- GZGG-2021-1/Seed Industry Research Project of Gansu Province
- GZGG-2021-1/Seed Industry Research Project of Gansu Province
- GZGG-2021-1/Seed Industry Research Project of Gansu Province
- GZGG-2021-1/Seed Industry Research Project of Gansu Province
- GZGG-2021-1/Seed Industry Research Project of Gansu Province
- GZGG-2021-1/Seed Industry Research Project of Gansu Province
- GZGG-2021-1/Seed Industry Research Project of Gansu Province
- GZGG-2021-1/Seed Industry Research Project of Gansu Province
- GZGG-2021-1/Seed Industry Research Project of Gansu Province
- GZGG-2021-1/Seed Industry Research Project of Gansu Province
- GZGG-2021-1/Seed Industry Research Project of Gansu Province
- GZGG-2021-1/Seed Industry Research Project of Gansu Province
LinkOut - more resources
Full Text Sources

