Adult fibroblasts use aggresomes only in distinct cell-states
- PMID: 36056070
- PMCID: PMC9440096
- DOI: 10.1038/s41598-022-19055-1
Adult fibroblasts use aggresomes only in distinct cell-states
Abstract
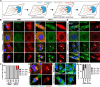
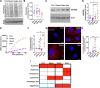
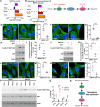
The aggresome is a protein turnover system in which proteins are trafficked along microtubules to the centrosome for degradation. Despite extensive focus on aggresomes in immortalized cell lines, it remains unclear if the aggresome is conserved in all primary cells and all cell-states. Here we examined the aggresome in primary adult mouse dermal fibroblasts shifted into four distinct cell-states. We found that in response to proteasome inhibition, quiescent and immortalized fibroblasts formed aggresomes, whereas proliferating and senescent fibroblasts did not. Using this model, we generated a resource to provide a characterization of the proteostasis networks in which the aggresome is used and transcriptomic features associated with the presence or absence of aggresome formation. Using this resource, we validate a previously reported role for p38 MAPK signaling in aggresome formation and identify TAK1 as a novel driver of aggresome formation upstream of p38 MAPKs. Together, our data demonstrate that the aggresome is a non-universal protein degradation system which can be used cell-state specifically and provide a resource for studying aggresome formation and function.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Formation of aggresomes with hydrogel-like characteristics by proteasome inhibition.Biochim Biophys Acta Gene Regul Mech. 2023 Jun;1866(2):194932. doi: 10.1016/j.bbagrm.2023.194932. Epub 2023 Mar 28. Biochim Biophys Acta Gene Regul Mech. 2023. PMID: 36997115
-
Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera.J Cell Biol. 1999 Sep 20;146(6):1239-54. doi: 10.1083/jcb.146.6.1239. J Cell Biol. 1999. PMID: 10491388 Free PMC article.
-
Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein.Hum Mol Genet. 2003 Apr 1;12(7):749-57. doi: 10.1093/hmg/ddg074. Hum Mol Genet. 2003. PMID: 12651870
-
Aggresome formation and neurodegenerative diseases: therapeutic implications.Curr Med Chem. 2008;15(1):47-60. doi: 10.2174/092986708783330692. Curr Med Chem. 2008. PMID: 18220762 Free PMC article. Review.
-
Aggresomes, inclusion bodies and protein aggregation.Trends Cell Biol. 2000 Dec;10(12):524-30. doi: 10.1016/s0962-8924(00)01852-3. Trends Cell Biol. 2000. PMID: 11121744 Review.
References
-
- Fusco C, Micale L, Egorov M, Monti M, D'Addetta EV, Augello B, Cozzolino F, Calcagni A, Fontana A, Polishchuk RS, Didelot G, Reymond A, Pucci P, Merla G. The E3-ubiquitin ligase TRIM50 interacts with HDAC6 and p62, and promotes the sequestration and clearance of ubiquitinated proteins into the aggresome. PLoS One. 2012;7(7):e40440. doi: 10.1371/journal.pone.0040440. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

