Aberrant astrocyte protein secretion contributes to altered neuronal development in multiple models of neurodevelopmental disorders
- PMID: 36042312
- PMCID: PMC10395413
- DOI: 10.1038/s41593-022-01150-1
Aberrant astrocyte protein secretion contributes to altered neuronal development in multiple models of neurodevelopmental disorders
Abstract
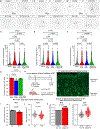
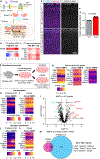
Astrocytes negatively impact neuronal development in many models of neurodevelopmental disorders (NDs); however, how they do this, and if mechanisms are shared across disorders, is not known. In this study, we developed a cell culture system to ask how astrocyte protein secretion and gene expression change in three mouse models of genetic NDs (Rett, Fragile X and Down syndromes). ND astrocytes increase release of Igfbp2, a secreted inhibitor of insulin-like growth factor (IGF). IGF rescues neuronal deficits in many NDs, and we found that blocking Igfbp2 partially rescues inhibitory effects of Rett syndrome astrocytes, suggesting that increased astrocyte Igfbp2 contributes to decreased IGF signaling in NDs. We identified that increased BMP signaling is upstream of protein secretion changes, including Igfbp2, and blocking BMP signaling in Fragile X and Rett syndrome astrocytes reverses inhibitory effects on neurite outgrowth. This work provides a resource of astrocyte-secreted proteins in health and ND models and identifies novel targets for intervention in diverse NDs.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing Interests
The authors declare no competing interests.
Figures








Similar articles
-
An Astrocytic Influence on Impaired Tonic Inhibition in Hippocampal CA1 Pyramidal Neurons in a Mouse Model of Rett Syndrome.J Neurosci. 2020 Aug 5;40(32):6250-6261. doi: 10.1523/JNEUROSCI.3042-19.2020. Epub 2020 Jul 2. J Neurosci. 2020. PMID: 32616668 Free PMC article.
-
Astrocyte Transcriptome from the Mecp2(308)-Truncated Mouse Model of Rett Syndrome.Neuromolecular Med. 2015 Dec;17(4):353-63. doi: 10.1007/s12017-015-8363-9. Epub 2015 Jul 25. Neuromolecular Med. 2015. PMID: 26208914
-
Activity-dependent modulation of synapse-regulating genes in astrocytes.Elife. 2021 Sep 8;10:e70514. doi: 10.7554/eLife.70514. Elife. 2021. PMID: 34494546 Free PMC article.
-
Deficits in olfactory system neurogenesis in neurodevelopmental disorders.Genesis. 2024 Apr;62(2):e23590. doi: 10.1002/dvg.23590. Genesis. 2024. PMID: 38490949 Review.
-
The application of human pluripotent stem cells to model the neuronal and glial components of neurodevelopmental disorders.Mol Psychiatry. 2020 Feb;25(2):368-378. doi: 10.1038/s41380-019-0495-0. Epub 2019 Aug 27. Mol Psychiatry. 2020. PMID: 31455859 Review.
Cited by
-
Metabolic dynamics in astrocytes and microglia during post-natal development and their implications for autism spectrum disorders.Front Cell Neurosci. 2024 Feb 14;18:1354259. doi: 10.3389/fncel.2024.1354259. eCollection 2024. Front Cell Neurosci. 2024. PMID: 38419654 Free PMC article. Review.
-
Astrocytic APOE4 genotype-mediated negative impacts on synaptic architecture in human pluripotent stem cell model.Stem Cell Reports. 2023 Sep 12;18(9):1854-1869. doi: 10.1016/j.stemcr.2023.08.002. Epub 2023 Aug 31. Stem Cell Reports. 2023. PMID: 37657448 Free PMC article.
-
The mTOR Pathway: A Common Link Between Alzheimer's Disease and Down Syndrome.J Clin Med. 2024 Oct 17;13(20):6183. doi: 10.3390/jcm13206183. J Clin Med. 2024. PMID: 39458132 Free PMC article. Review.
-
Brain-specific lipoprotein receptors interact with astrocyte derived apolipoprotein and mediate neuron-glia lipid shuttling.Nat Commun. 2021 Apr 23;12(1):2408. doi: 10.1038/s41467-021-22751-7. Nat Commun. 2021. PMID: 33893307 Free PMC article.
-
Astrocyte-secreted neurocan controls inhibitory synapse formation and function.Neuron. 2024 May 15;112(10):1657-1675.e10. doi: 10.1016/j.neuron.2024.03.007. Epub 2024 Apr 3. Neuron. 2024. PMID: 38574730
References
-
- Ebrahimi-Fakhari D.& Sahin M.Autism and the synapse: emerging mechanisms and mechanism-based therapies. Current opinion in neurology 28, 91–102 (2015). - PubMed
-
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U.& Zoghbi HY Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nature genetics 23, 185–188 (1999). - PubMed
-
- Irwin SA, Galvez R.& Greenough WT Dendritic Spine Structural Anomalies in Fragile-X Mental Retardation Syndrome. (2000). - PubMed
Methods References
-
- Guy J, Hendrich B, Holmes M, Martin JE & Bird A.A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nature genetics 27, 322–326 (2001). - PubMed
-
- Bakker CE, Verheij C, Willemsen R, van der Helm R, Oerlemans F, Vermey M, Bygrave A, Hoogeveen A, Oostra BA, Reyniers E, De Boule K, D’Hooge R, Cras P, van Velzen D, Nagels G, Martin J-J, De Deyn PP, Darby JK & Willems PJ Fmr1 knockout mice: A model to study fragile X mental retardation. Cell 78, 23–33 (1994). - PubMed
-
- Davisson MT, Schmidt C.& Akeson EC Segmental trisomy of murine chromosome 16: a new model system for studying Down syndrome. Progress in clinical and biological research 360, 263–280 (1990). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

