Pan-Cancer Analysis of Canonical and Modified miRNAs Enhances the Resolution of the Functional miRNAome in Cancer
- PMID: 36040379
- PMCID: PMC9574374
- DOI: 10.1158/0008-5472.CAN-22-0240
Pan-Cancer Analysis of Canonical and Modified miRNAs Enhances the Resolution of the Functional miRNAome in Cancer
Abstract
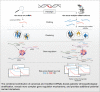
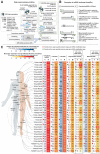
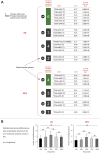
Epitranscriptomic studies of miRNAs have added a new layer of complexity to the cancer field. Although there is fast-growing interest in adenosine-to-inosine (A-to-I) miRNA editing and alternative cleavage that shifts miRNA isoforms, simultaneous evaluation of both modifications in cancer is still missing. Here, we concurrently profiled multiple miRNA modification types, including A-to-I miRNA editing and shifted miRNA isoforms, in >13,000 adult and pediatric tumor samples across 38 distinct cancer cohorts from The Cancer Genome Atlas and The Therapeutically Applicable Research to Generate Effective Treatments data sets. The differences between canonical miRNAs and the wider miRNAome in terms of expression, clustering, dysregulation, and prognostic standpoint were investigated. The combination of canonical miRNAs and modified miRNAs boosted the quality of clustering results, outlining unique clinicopathologic features among cohorts. Certain modified miRNAs showed opposite expression from their canonical counterparts in cancer, potentially impacting their targets and function. Finally, a shifted and edited miRNA isoform was experimentally validated to directly bind and suppress a unique target. These findings outline the importance of going beyond the well-established paradigm of one mature miRNA per miRNA arm to elucidate novel mechanisms related to cancer progression.
Significance: Modified miRNAs may act as cancer biomarkers and function as allies or antagonists of their canonical counterparts in gene regulation, suggesting the concurrent consideration of canonical and modified miRNAs can boost patient stratification.
©2022 The Authors; Published by the American Association for Cancer Research.
Figures



Similar articles
-
Human cancer tissues exhibit reduced A-to-I editing of miRNAs coupled with elevated editing of their targets.Nucleic Acids Res. 2018 Jan 9;46(1):71-82. doi: 10.1093/nar/gkx1176. Nucleic Acids Res. 2018. PMID: 29165639 Free PMC article.
-
Systematic characterization of A-to-I RNA editing hotspots in microRNAs across human cancers.Genome Res. 2017 Jul;27(7):1112-1125. doi: 10.1101/gr.219741.116. Epub 2017 Apr 14. Genome Res. 2017. PMID: 28411194 Free PMC article.
-
RNA editing of human microRNAs.Genome Biol. 2006;7(4):R27. doi: 10.1186/gb-2006-7-4-r27. Epub 2006 Apr 4. Genome Biol. 2006. PMID: 16594986 Free PMC article.
-
When MicroRNAs Meet RNA Editing in Cancer: A Nucleotide Change Can Make a Difference.Bioessays. 2018 Feb;40(2):10.1002/bies.201700188. doi: 10.1002/bies.201700188. Epub 2017 Dec 27. Bioessays. 2018. PMID: 29280160 Free PMC article. Review.
-
The adaptive potential of RNA editing-mediated miRNA-retargeting in cancer.Biochim Biophys Acta Gene Regul Mech. 2019 Mar;1862(3):291-300. doi: 10.1016/j.bbagrm.2018.12.007. Epub 2018 Dec 31. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 30605729 Review.
Cited by
-
Functional and Potential Therapeutic Implication of MicroRNAs in Pancreatic Cancer.Int J Mol Sci. 2023 Dec 15;24(24):17523. doi: 10.3390/ijms242417523. Int J Mol Sci. 2023. PMID: 38139352 Free PMC article. Review.
-
Poly(A)-specific RNase (PARN) generates and regulates miR-125a-5p 3'-isoforms, displaying an altered expression in breast cancer.Signal Transduct Target Ther. 2024 Apr 15;9(1):90. doi: 10.1038/s41392-024-01795-3. Signal Transduct Target Ther. 2024. PMID: 38616203 Free PMC article. No abstract available.
-
tRFUniverse: A comprehensive resource for the interactive analyses of tRNA-derived ncRNAs in human cancer.iScience. 2024 Jan 5;27(2):108810. doi: 10.1016/j.isci.2024.108810. eCollection 2024 Feb 16. iScience. 2024. PMID: 38303722 Free PMC article.
-
Improved lung cancer classification by employing diverse molecular features of microRNAs.Heliyon. 2024 Feb 10;10(4):e26081. doi: 10.1016/j.heliyon.2024.e26081. eCollection 2024 Feb 29. Heliyon. 2024. PMID: 38384512 Free PMC article.
-
Reassessment of miRNA variant (isomiRs) composition by small RNA sequencing.Cell Rep Methods. 2023 May 16;3(5):100480. doi: 10.1016/j.crmeth.2023.100480. eCollection 2023 May 22. Cell Rep Methods. 2023. PMID: 37323569 Free PMC article.
References
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–97. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

