Annotations of novel antennae-expressed genes in male Glossina morsitans morsitans tsetse flies
- PMID: 36037171
- PMCID: PMC9423656
- DOI: 10.1371/journal.pone.0273543
Annotations of novel antennae-expressed genes in male Glossina morsitans morsitans tsetse flies
Abstract
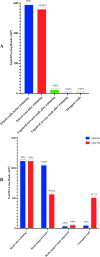
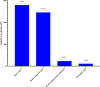
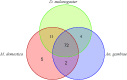
Tsetse flies use antennal expressed genes to navigate their environment. While most canonical genes associated with chemoreception are annotated, potential gaps with important antennal genes are uncharacterized in Glossina morsitans morsitans. We generated antennae-specific transcriptomes from adult male G. m. morsitans flies fed/unfed on bloodmeal and/or exposed to an attractant (ε-nonalactone), a repellant (δ-nonalactone) or paraffin diluent. Using bioinformatics approach, we mapped raw reads onto G. m. morsitans gene-set from VectorBase and collected un-mapped reads (constituting the gaps in annotation). We de novo assembled these reads (un-mapped) into transcript and identified corresponding genes of the transcripts in G. m. morsitans gene-set and protein homologs in UniProt protein database to further annotate the gaps. We predicted potential protein-coding gene regions associated with these transcripts in G. m. morsitans genome, annotated/curated these genes and identified their putative annotated orthologs/homologs in Drosophila melanogaster, Musca domestica or Anopheles gambiae genomes. We finally evaluated differential expression of the novel genes in relation to odor exposures relative to no-odor control (unfed flies). About 45.21% of the sequenced reads had no corresponding transcripts within G. m. morsitans gene-set, corresponding to the gap in existing annotation of the tsetse fly genome. The total reads assembled into 72,428 unique transcripts, most (74.43%) of which had no corresponding genes in the UniProt database. We annotated/curated 592 genes from these transcripts, among which 202 were novel while 390 were improvements of existing genes in the G. m. morsitans genome. Among the novel genes, 94 had orthologs in D. melanogaster, M. domestica or An. gambiae while 88 had homologs in UniProt. These orthologs were putatively associated with oxidative regulation, protein synthesis, transcriptional and/or translational regulation, detoxification and metal ion binding, thus providing insight into their specific roles in antennal physiological processes in male G. m. morsitans. A novel gene (GMOY014237.R1396) was differentially expressed in response to the attractant. We thus established significant gaps in G. m. morsitans genome annotation and identified novel male antennae-expressed genes in the genome, among which > 53% (108) are potentially G. m. morsitans specific.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Expansions of chemosensory gene orthologs among selected tsetse fly species and their expressions in Glossina morsitans morsitans tsetse fly.PLoS Negl Trop Dis. 2020 Jun 26;14(6):e0008341. doi: 10.1371/journal.pntd.0008341. eCollection 2020 Jun. PLoS Negl Trop Dis. 2020. PMID: 32589659 Free PMC article.
-
Odorant and gustatory receptors in the tsetse fly Glossina morsitans morsitans.PLoS Negl Trop Dis. 2014 Apr 24;8(4):e2663. doi: 10.1371/journal.pntd.0002663. eCollection 2014 Apr. PLoS Negl Trop Dis. 2014. PMID: 24763191 Free PMC article.
-
Expression Levels of Odorant Receptor Genes in the Savanna Tsetse Fly, Glossina morsitans morsitans.J Med Entomol. 2018 Jun 28;55(4):855-861. doi: 10.1093/jme/tjy018. J Med Entomol. 2018. PMID: 29529232
-
Genome-Wide Comparative Analysis of Chemosensory Gene Families in Five Tsetse Fly Species.PLoS Negl Trop Dis. 2016 Feb 17;10(2):e0004421. doi: 10.1371/journal.pntd.0004421. eCollection 2016 Feb. PLoS Negl Trop Dis. 2016. PMID: 26886411 Free PMC article.
-
Responses of Glossina pallidipes and Glossina morsitans morsitans tsetse flies to analogues of δ-octalactone and selected blends.Acta Trop. 2016 Aug;160:53-7. doi: 10.1016/j.actatropica.2016.04.011. Epub 2016 Apr 30. Acta Trop. 2016. PMID: 27143219 Free PMC article.
References
-
- Vale GA. The responses of tsetse flies (Diptera, Glossinidae) to mobile and stationary baits. Bull. Entomol. Res. 1974. Dec; 64 (4); 545–588. doi: 10.1017/S0007485300035860 - DOI
-
- Vale GA. Field studies of the responses of tsetse flies (Glossinidae) and other Diptera to carbon dioxide, acetone and other chemicals. Bull. Entomol. Res. 1980. Dec; 70 (4); 563–570. doi: 10.1017/S0007485300007860 - DOI
-
- Vale GA, Hall DR, Gough AJE. The olfactory responses of tsetse flies, Glossina spp. (Diptera: Glossinidae), to phenols and urine in the field. Bull. Entomol. Res. 1988. Dec; 78(2); 293–300. doi: 10.1017/s0007485300013055 - DOI
-
- Gikonyo NK, Hassanali A, Njagi PG, Saini RK. Behaviour of Glossina morsitans morsitans Westwood (Diptera: Glossinidae) on waterbuck Kobus defassa Ruppel and feeding membranes smeared with waterbuck sebum indicates the presence of allomones. Acta Trop. 2000. Dec 1;77(3):295–303. doi: 10.1016/s0001-706x(00)00153-4 - DOI - PubMed
-
- Gikonyo NK, Hassanali A, Njagi PG, Saini RK. Responses of Glossina morsitans morsitans to blends of electroantennographically active compounds in the odors of its preferred (buffalo and ox) and nonpreferred (waterbuck) hosts. J Chem Ecol. 2003. Oct;29(10):2331–45. doi: 10.1023/a:1026230615877 - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

