IκBζ controls IL-17-triggered gene expression program in intestinal epithelial cells that restricts colonization of SFB and prevents Th17-associated pathologies
- PMID: 35999460
- PMCID: PMC9705257
- DOI: 10.1038/s41385-022-00554-3
IκBζ controls IL-17-triggered gene expression program in intestinal epithelial cells that restricts colonization of SFB and prevents Th17-associated pathologies
Abstract
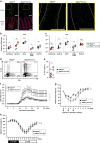
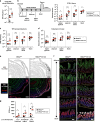
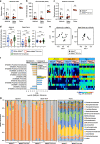
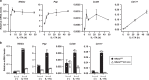
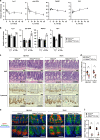
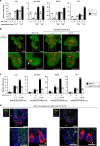
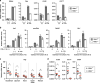
Control of gut microbes is crucial for not only local defense in the intestine but also proper systemic immune responses. Although intestinal epithelial cells (IECs) play important roles in cytokine-mediated control of enterobacteria, the underlying mechanisms are not fully understood. Here we show that deletion of IκBζ in IECs in mice leads to dysbiosis with marked expansion of segmented filamentous bacteria (SFB), thereby enhancing Th17 cell development and exacerbating inflammatory diseases. Mechanistically, the IκBζ deficiency results in decrease in the number of Paneth cells and impairment in expression of IL-17-inducible genes involved in IgA production. The decrease in Paneth cells is caused by aberrant activation of IFN-γ signaling and a failure of IL-17-dependent recovery from IFN-γ-induced damage. Thus, the IL-17R-IκBζ axis in IECs contributes to the maintenance of intestinal homeostasis by serving as a key component in a regulatory loop between the gut microbiota and immune cells.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Induction of Intestinal Th17 Cells by Flagellins From Segmented Filamentous Bacteria.Front Immunol. 2019 Nov 22;10:2750. doi: 10.3389/fimmu.2019.02750. eCollection 2019. Front Immunol. 2019. PMID: 31824516 Free PMC article.
-
Intestinal Interleukin-17 Receptor Signaling Mediates Reciprocal Control of the Gut Microbiota and Autoimmune Inflammation.Immunity. 2016 Mar 15;44(3):659-671. doi: 10.1016/j.immuni.2016.02.007. Immunity. 2016. PMID: 26982366 Free PMC article.
-
Deficiency in intestinal epithelial O-GlcNAcylation predisposes to gut inflammation.EMBO Mol Med. 2018 Aug;10(8):e8736. doi: 10.15252/emmm.201708736. EMBO Mol Med. 2018. PMID: 29941542 Free PMC article.
-
The effect of probiotics and gut microbiota on Th17 cells.Int Rev Immunol. 2013 Oct-Dec;32(5-6):511-25. doi: 10.3109/08830185.2013.839665. Epub 2013 Oct 4. Int Rev Immunol. 2013. PMID: 24094077 Review.
-
Development, validation and implementation of an in vitro model for the study of metabolic and immune function in normal and inflamed human colonic epithelium.Dan Med J. 2015 Jan;62(1):B4973. Dan Med J. 2015. PMID: 25557335 Review.
Cited by
-
The Nuclear NF-κB Regulator IκBζ: Updates on Its Molecular Functions and Pathophysiological Roles.Cells. 2024 Aug 31;13(17):1467. doi: 10.3390/cells13171467. Cells. 2024. PMID: 39273036 Free PMC article. Review.
-
IκBζ is an essential mediator of immunity to oropharyngeal candidiasis.Cell Host Microbe. 2023 Oct 11;31(10):1700-1713.e4. doi: 10.1016/j.chom.2023.08.016. Epub 2023 Sep 18. Cell Host Microbe. 2023. PMID: 37725983 Free PMC article.
-
The central inflammatory regulator IκBζ: induction, regulation and physiological functions.Front Immunol. 2023 Jun 12;14:1188253. doi: 10.3389/fimmu.2023.1188253. eCollection 2023. Front Immunol. 2023. PMID: 37377955 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

