Intestinal single-cell atlas reveals novel lymphocytes in pigs with similarities to human cells
- PMID: 35995567
- PMCID: PMC9396248
- DOI: 10.26508/lsa.202201442
Intestinal single-cell atlas reveals novel lymphocytes in pigs with similarities to human cells
Abstract
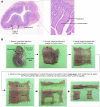
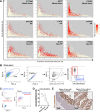
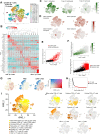
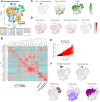
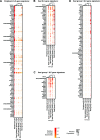
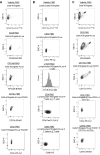
Lymphocytes can heavily influence intestinal health, but resolving intestinal lymphocyte function is challenging as the intestine contains a vastly heterogeneous mixture of cells. Pigs are an advantageous biomedical model, but deeper understanding of intestinal lymphocytes is warranted to improve model utility. Twenty-six cell types were identified in the porcine ileum by single-cell RNA sequencing and further compared with cells in human and murine ileum. Though general consensus of cell subsets across species was revealed, some porcine-specific lymphocyte subsets were identified. Differential tissue dissection and in situ analyses conferred spatial context, revealing similar locations of lymphocyte subsets in Peyer's patches and epithelium in pig-to-human comparisons. Like humans, activated and effector lymphocytes were abundant in the ileum but not periphery of pigs, suggesting tissue-specific and/or activation-associated gene expression. Gene signatures for peripheral and ileal innate lymphoid cells newly discovered in pigs were defined and highlighted similarities to human innate lymphoid cells. Overall, we reveal novel lymphocyte subsets in pigs and highlight utility of pigs for intestinal research applications.
© 2022 Wiarda et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures














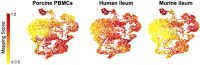












Similar articles
-
Characterization of autonomic nerve markers and lymphocyte subsets in the ileal Peyer's patch of pigs infected experimentally with Brachyspira hyodysenteriae.J Comp Pathol. 2010 Nov;143(4):248-57. doi: 10.1016/j.jcpa.2010.04.003. Epub 2010 Jun 4. J Comp Pathol. 2010. PMID: 20605161
-
Immunome differences between porcine ileal and jejunal Peyer's patches revealed by global transcriptome sequencing of gut-associated lymphoid tissues.Sci Rep. 2018 Jun 13;8(1):9077. doi: 10.1038/s41598-018-27019-7. Sci Rep. 2018. PMID: 29899562 Free PMC article.
-
Development of the natural response of immunoglobulin secreting cells in the pig as a function of organ, age and housing.Dev Comp Immunol. 1999 Sep;23(6):511-20. doi: 10.1016/s0145-305x(99)00026-9. Dev Comp Immunol. 1999. PMID: 10512461
-
The intestinal habitat for organized lymphoid tissues in ruminants; comparative aspects of structure, function and development.Vet Immunol Immunopathol. 1991 Mar;28(1):1-16. doi: 10.1016/0165-2427(91)90038-e. Vet Immunol Immunopathol. 1991. PMID: 1905075 Review.
-
Intraepithelial lymphocytes in the pig intestine: T cell and innate lymphoid cell contributions to intestinal barrier immunity.Front Immunol. 2022 Dec 8;13:1048708. doi: 10.3389/fimmu.2022.1048708. eCollection 2022. Front Immunol. 2022. PMID: 36569897 Free PMC article. Review.
Cited by
-
Comparative transcriptome analysis of T lymphocyte subpopulations and identification of critical regulators defining porcine thymocyte identity.Front Immunol. 2024 Feb 7;15:1339787. doi: 10.3389/fimmu.2024.1339787. eCollection 2024. Front Immunol. 2024. PMID: 38384475 Free PMC article.
-
Regional epithelial cell diversity in the small intestine of pigs.J Anim Sci. 2023 Jan 3;101:skac318. doi: 10.1093/jas/skac318. J Anim Sci. 2023. PMID: 36183288 Free PMC article.
-
Alternatives to antibiotics in pig production: looking through the lens of immunophysiology.Stress Biol. 2024 Jan 2;4(1):1. doi: 10.1007/s44154-023-00134-w. Stress Biol. 2024. PMID: 38163818 Free PMC article. Review.
-
The formidable guardian: Type 3 immunity in the intestine of pigs.Virulence. 2024 Dec;15(1):2424325. doi: 10.1080/21505594.2024.2424325. Epub 2024 Nov 8. Virulence. 2024. PMID: 39497434 Free PMC article. Review.
-
Advances in single-cell transcriptomics in animal research.J Anim Sci Biotechnol. 2024 Aug 2;15(1):102. doi: 10.1186/s40104-024-01063-y. J Anim Sci Biotechnol. 2024. PMID: 39090689 Free PMC article. Review.
References
-
- Annamalai T, Lu Z, Jung K, Langel SN, Tuggle CK, Dekkers JCM, Waide EH, Kandasamy S, Saif LJ (2019) Infectivity of GII.4 human norovirus does not differ between T-B-NK+ severe combined immunodeficiency (SCID) and non-SCID gnotobiotic pigs, implicating the role of NK cells in mediation of human norovirus infection. Virus Res 267: 21–25. 10.1016/j.virusres.2019.05.002 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
