A pendulum of induction between the epiblast and extra-embryonic endoderm supports post-implantation progression
- PMID: 35993866
- PMCID: PMC9534490
- DOI: 10.1242/dev.192310
A pendulum of induction between the epiblast and extra-embryonic endoderm supports post-implantation progression
Abstract
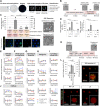
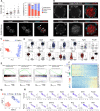
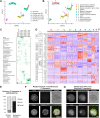
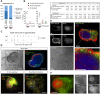
Embryogenesis is supported by dynamic loops of cellular interactions. Here, we create a partial mouse embryo model to elucidate the principles of epiblast (Epi) and extra-embryonic endoderm co-development (XEn). We trigger naive mouse embryonic stem cells to form a blastocyst-stage niche of Epi-like cells and XEn-like cells (3D, hydrogel free and serum free). Once established, these two lineages autonomously progress in minimal medium to form an inner pro-amniotic-like cavity surrounded by polarized Epi-like cells covered with visceral endoderm (VE)-like cells. The progression occurs through reciprocal inductions by which the Epi supports the primitive endoderm (PrE) to produce a basal lamina that subsequently regulates Epi polarization and/or cavitation, which, in return, channels the transcriptomic progression to VE. This VE then contributes to Epi bifurcation into anterior- and posterior-like states. Similarly, boosting the formation of PrE-like cells within blastoids supports developmental progression. We argue that self-organization can arise from lineage bifurcation followed by a pendulum of induction that propagates over time.
Keywords: Blastoids; Embryonic stem cells; Extra-embryonic endoderm/epiblast rosette; Post-implantation development; Primitive endoderm; Pro-amniotic cavity.
© 2022. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests N.C.R., E.J.V. and C.A.v.B. are inventors on the patents US14/784,659 and PCT/NL2014/050239, which describe the formation of mouse blastoids (April 2014). All rights and duties are maintained by the Institute for Molecular Biotechnology, Austrian Academy of Science.
Figures



Similar articles
-
Journey of the mouse primitive endoderm: from specification to maturation.Philos Trans R Soc Lond B Biol Sci. 2022 Dec 5;377(1865):20210252. doi: 10.1098/rstb.2021.0252. Epub 2022 Oct 17. Philos Trans R Soc Lond B Biol Sci. 2022. PMID: 36252215 Free PMC article. Review.
-
Self-Organization of Mouse Stem Cells into an Extended Potential Blastoid.Dev Cell. 2019 Dec 16;51(6):698-712.e8. doi: 10.1016/j.devcel.2019.11.014. Dev Cell. 2019. PMID: 31846649 Free PMC article.
-
Does mouse embryo primordial germ cell activation start before implantation as suggested by single-cell transcriptomics dynamics?Mol Hum Reprod. 2016 Mar;22(3):208-25. doi: 10.1093/molehr/gav072. Epub 2016 Jan 5. Mol Hum Reprod. 2016. PMID: 26740066
-
A comparative analysis of extra-embryonic endoderm cell lines.PLoS One. 2010 Aug 6;5(8):e12016. doi: 10.1371/journal.pone.0012016. PLoS One. 2010. PMID: 20711519 Free PMC article.
-
[Epiblast and primitive endoderm cell specification during mouse preimplantation development: a combination between biology and mathematical modeling].Med Sci (Paris). 2016 Feb;32(2):192-7. doi: 10.1051/medsci/20163202013. Epub 2016 Mar 2. Med Sci (Paris). 2016. PMID: 26936177 Review. French.
Cited by
-
A comprehensive review: synergizing stem cell and embryonic development knowledge in mouse and human integrated stem cell-based embryo models.Front Cell Dev Biol. 2024 Apr 22;12:1386739. doi: 10.3389/fcell.2024.1386739. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38715920 Free PMC article. Review.
-
Topical section: embryonic models (2023) for Current Opinion in Genetics & Development.Curr Opin Genet Dev. 2024 Feb;84:102134. doi: 10.1016/j.gde.2023.102134. Epub 2023 Dec 5. Curr Opin Genet Dev. 2024. PMID: 38052116 Free PMC article. Review.
-
Shifting early embryology paradigms: Applications of stem cell-based embryo models in bioengineering.Curr Opin Genet Dev. 2023 Aug;81:102069. doi: 10.1016/j.gde.2023.102069. Epub 2023 Jun 29. Curr Opin Genet Dev. 2023. PMID: 37392541 Free PMC article. Review.
-
Technical challenges of studying early human development.Development. 2023 Jun 1;150(11):dev201797. doi: 10.1242/dev.201797. Epub 2023 Jun 1. Development. 2023. PMID: 37260362 Free PMC article.
-
The logic of monsters: development and morphological diversity in stem-cell-based embryo models.Interface Focus. 2024 Oct 25;14(5):20240023. doi: 10.1098/rsfs.2024.0023. eCollection 2024 Oct 11. Interface Focus. 2024. PMID: 39464644 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

