Exploring whole proteome to contrive multi-epitope-based vaccine for NeoCoV: An immunoinformtics and in-silico approach
- PMID: 35990651
- PMCID: PMC9382669
- DOI: 10.3389/fimmu.2022.956776
Exploring whole proteome to contrive multi-epitope-based vaccine for NeoCoV: An immunoinformtics and in-silico approach
Abstract
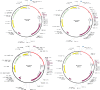
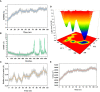
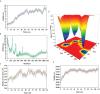
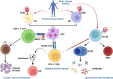
Neo-Coronavirus (NeoCoV) is a novel Betacoronavirus (β-CoVs or Beta-CoVs) discovered in bat specimens in South Africa during 2011. The viral sequence is highly similar to Middle East Respiratory Syndrome, particularly that of structural proteins. Thus, scientists have emphasized the threat posed by NeoCoV associated with human angiotensin-converting enzyme 2 (ACE2) usage, which could lead to a high death rate and faster transmission rate in humans. The development of a NeoCoV vaccine could provide a promising option for the future control of the virus in case of human infection. In silico predictions can decrease the number of experiments required, making the immunoinformatics approaches cost-effective and convenient. Herein, with the aid of immunoinformatics and reverse vaccinology, we aimed to formulate a multi-epitope vaccine that may be used to prevent and treat NeoCoV infection. Based on the NeoCoV proteins, B-cell, cytotoxic T lymphocyte (CTL), and helper T lymphocyte (HTL) epitopes were shortlisted. Four vaccines (Neo-1-4) were devised by fusing shortlisted epitopes with appropriate adjuvants and linkers. The secondary and three-dimensional structures of final vaccines were then predicted. The binding interactions of these potential vaccines with toll-like immune receptors (TLR-2, TLR-3, and TLR-4) and major histocompatibility complex molecules (MHC-I and II) reveal that they properly fit into the receptors' binding domains. Besides, Neo-1 and Neo-4 vaccines exhibited better docking energies of -101.08 kcal/mol and -114.47 kcal/mol, respectively, with TLR-3 as compared to other vaccine constructs. The constructed vaccines are highly antigenic, non-allergenic, soluble, non-toxic, and topologically assessable with good physiochemical characteristics. Codon optimization and in-silico cloning confirmed efficient expression of the designed vaccines in Escherichia coli strain K12. In-silico immune simulation indicated that Neo-1 and Neo-4 vaccines could induce a strong immune response against NeoCoV. Lastly, the binding stability and strong binding affinity of Neo-1 and Neo-4 with TLR-3 receptor were validated using molecular dynamics simulations and free energy calculations (Molecular Mechanics/Generalized Born Surface Area method). The final vaccines require experimental validation to establish their safety and effectiveness in preventing NeoCoV infections.
Keywords: NeoCoV; epitopes prediction; immunoinformatics; multi-epitope vaccine; subunit vaccine; vaccine design.
Copyright © 2022 Aziz, Waqas, Halim, Ali, Iqbal, Iqbal, Khan and Al-Harrasi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Immunoinformatics design of a novel epitope-based vaccine candidate against dengue virus.Sci Rep. 2021 Oct 5;11(1):19707. doi: 10.1038/s41598-021-99227-7. Sci Rep. 2021. PMID: 34611250 Free PMC article.
-
Contriving Multi-Epitope Subunit of Vaccine for COVID-19: Immunoinformatics Approaches.Front Immunol. 2020 Jul 28;11:1784. doi: 10.3389/fimmu.2020.01784. eCollection 2020. Front Immunol. 2020. PMID: 32849643 Free PMC article.
-
Design of a multi-epitope vaccine against six Nocardia species based on reverse vaccinology combined with immunoinformatics.Front Immunol. 2023 Feb 2;14:1100188. doi: 10.3389/fimmu.2023.1100188. eCollection 2023. Front Immunol. 2023. PMID: 36845087 Free PMC article.
-
Immunogenic multi-epitope-based vaccine development to combat cyclosporiasis of immunocompromised patients applying computational biology method.Exp Parasitol. 2023 May;248:108497. doi: 10.1016/j.exppara.2023.108497. Epub 2023 Mar 10. Exp Parasitol. 2023. PMID: 36906252 Review.
-
Multi-epitopevaccines, from design to expression; an in silico approach.Hum Immunol. 2024 May;85(3):110804. doi: 10.1016/j.humimm.2024.110804. Epub 2024 Apr 23. Hum Immunol. 2024. PMID: 38658216 Review.
Cited by
-
Computational design and evaluation of mRNA- and protein-based conjugate vaccines for influenza A and SARS-CoV-2 viruses.J Genet Eng Biotechnol. 2023 Nov 15;21(1):120. doi: 10.1186/s43141-023-00574-x. J Genet Eng Biotechnol. 2023. PMID: 37966525 Free PMC article.
-
Virus-like particle vaccines with epitopes from porcine epidemic virus and transmissible gastroenteritis virus incorporated into self-assembling ADDomer platform provide clinical immune responses in piglets.Front Immunol. 2023 Oct 24;14:1251001. doi: 10.3389/fimmu.2023.1251001. eCollection 2023. Front Immunol. 2023. PMID: 37942329 Free PMC article.
-
Recent Advances in Studying Toll-like Receptors with the Use of Computational Methods.J Chem Inf Model. 2023 Jun 26;63(12):3669-3687. doi: 10.1021/acs.jcim.3c00419. Epub 2023 Jun 7. J Chem Inf Model. 2023. PMID: 37285179 Free PMC article. Review.
-
Development of multi-epitope vaccines against the monkeypox virus based on envelope proteins using immunoinformatics approaches.Front Immunol. 2023 Mar 13;14:1112816. doi: 10.3389/fimmu.2023.1112816. eCollection 2023. Front Immunol. 2023. PMID: 36993967 Free PMC article.
-
Contriving multi-epitope vaccine ensemble for monkeypox disease using an immunoinformatics approach.Front Immunol. 2022 Oct 13;13:1004804. doi: 10.3389/fimmu.2022.1004804. eCollection 2022. Front Immunol. 2022. PMID: 36311762 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

