The role of mtDNA in oocyte quality and embryo development
- PMID: 35986715
- PMCID: PMC10952685
- DOI: 10.1002/mrd.23640
The role of mtDNA in oocyte quality and embryo development
Abstract
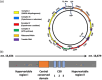
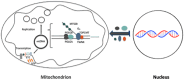
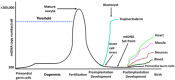
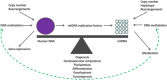
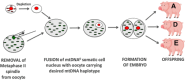
The mitochondrial genome resides in the mitochondria present in nearly all cell types. The porcine (Sus scrofa) mitochondrial genome is circa 16.7 kb in size and exists in the multimeric format in cells. Individual cell types have different numbers of mitochondrial DNA (mtDNA) copy number based on their requirements for ATP produced by oxidative phosphorylation. The oocyte has the largest number of mtDNA of any cell type. During oogenesis, the oocyte sets mtDNA copy number in order that sufficient copies are available to support subsequent developmental events. It also initiates a program of epigenetic patterning that regulates, for example, DNA methylation levels of the nuclear genome. Once fertilized, the nuclear and mitochondrial genomes establish synchrony to ensure that the embryo and fetus can complete each developmental milestone. However, altering the oocyte's mtDNA copy number by mitochondrial supplementation can affect the programming and gene expression profiles of the developing embryo and, in oocytes deficient of mtDNA, it appears to have a positive impact on the embryo development rates and gene expression profiles. Furthermore, mtDNA haplotypes, which define common maternal origins, appear to affect developmental outcomes and certain reproductive traits. Nevertheless, the manipulation of the mitochondrial content of an oocyte might have a developmental advantage.
Keywords: genomic balance; mitochondrial DNA; mitochondrial supplementation; nuclear transfer; oogenesis.
© 2022 The Authors. Molecular Reproduction and Development published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Mitochondrial supplementation of Sus scrofa metaphase II oocytes alters DNA methylation and gene expression profiles of blastocysts.Epigenetics Chromatin. 2022 Apr 15;15(1):12. doi: 10.1186/s13072-022-00442-x. Epigenetics Chromatin. 2022. PMID: 35428319 Free PMC article.
-
Additional mitochondrial DNA influences the interactions between the nuclear and mitochondrial genomes in a bovine embryo model of nuclear transfer.Sci Rep. 2018 May 8;8(1):7246. doi: 10.1038/s41598-018-25516-3. Sci Rep. 2018. PMID: 29740154 Free PMC article.
-
Use of Bisection to Reduce Mitochondrial DNA in the Bovine Oocyte.J Vis Exp. 2022 Jul 6;(185). doi: 10.3791/64060. J Vis Exp. 2022. PMID: 35876541
-
Mitochondrial DNA Assessment to Determine Oocyte and Embryo Viability.Semin Reprod Med. 2015 Nov;33(6):401-9. doi: 10.1055/s-0035-1567821. Epub 2015 Nov 13. Semin Reprod Med. 2015. PMID: 26565384 Review.
-
Mitochondrial function in the human oocyte and embryo and their role in developmental competence.Mitochondrion. 2011 Sep;11(5):797-813. doi: 10.1016/j.mito.2010.09.012. Epub 2010 Oct 7. Mitochondrion. 2011. PMID: 20933103 Review.
Cited by
-
Dynamics of Mitochondrial DNA Copy Number and Membrane Potential in Mouse Pre-Implantation Embryos: Responses to Diverse Types of Oxidative Stress.Genes (Basel). 2024 Mar 16;15(3):367. doi: 10.3390/genes15030367. Genes (Basel). 2024. PMID: 38540426 Free PMC article.
-
Mitochondrial DNA Supplementation of Oocytes Has Downstream Effects on the Transcriptional Profiles of Sus scrofa Adult Tissues with High mtDNA Copy Number.Int J Mol Sci. 2023 Apr 19;24(8):7545. doi: 10.3390/ijms24087545. Int J Mol Sci. 2023. PMID: 37108708 Free PMC article.
-
Overview of Gene Expression Dynamics during Human Oogenesis/Folliculogenesis.Int J Mol Sci. 2023 Dec 19;25(1):33. doi: 10.3390/ijms25010033. Int J Mol Sci. 2023. PMID: 38203203 Free PMC article. Review.
-
Mitochondrial DNA Deficiency and Supplementation in Sus scrofa Oocytes Influence Transcriptome Profiles in Oocytes and Blastocysts.Int J Mol Sci. 2023 Feb 14;24(4):3783. doi: 10.3390/ijms24043783. Int J Mol Sci. 2023. PMID: 36835193 Free PMC article.
-
Aging-Related Ovarian Failure and Infertility: Melatonin to the Rescue.Antioxidants (Basel). 2023 Mar 11;12(3):695. doi: 10.3390/antiox12030695. Antioxidants (Basel). 2023. PMID: 36978942 Free PMC article. Review.
References
-
- Anderson, S. , Bankier, A. T. , Barrell, B. G. , de Bruijn, M. H. , Coulson, A. R. , Drouin, J. , Eperon, I. C. , Nierlich, D. P. , Roe, B. A. , Sanger, F. , Schreier, P. H. , Smith, A. J. , Staden, R. , & Young, I. G. (1981). Sequence and organization of the human mitochondrial genome. Nature, 290(5806), 457–465. http://www.ncbi.nlm.nih.gov/pubmed/7219534 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

