The endogenous HBZ interactome in ATL leukemic cells reveals an unprecedented complexity of host interacting partners involved in RNA splicing
- PMID: 35979358
- PMCID: PMC9376625
- DOI: 10.3389/fimmu.2022.939863
The endogenous HBZ interactome in ATL leukemic cells reveals an unprecedented complexity of host interacting partners involved in RNA splicing
Abstract
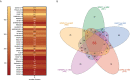
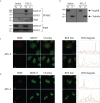
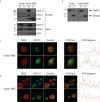
Adult T-cell leukemia/lymphoma (ATL) is a T-cell lymphoproliferative neoplasm caused by the human T-cell leukemia virus type 1 (HTLV-1). Two viral proteins, Tax-1 and HBZ play important roles in HTLV-1 infectivity and in HTLV-1-associated pathologies by altering key pathways of cell homeostasis. However, the molecular mechanisms through which the two viral proteins, particularly HBZ, induce and/or sustain the oncogenic process are still largely elusive. Previous results suggested that HBZ interaction with nuclear factors may alter cell cycle and cell proliferation. To have a more complete picture of the HBZ interactions, we investigated in detail the endogenous HBZ interactome in leukemic cells by immunoprecipitating the HBZ-interacting complexes of ATL-2 leukemic cells, followed by tandem mass spectrometry analyses. RNA seq analysis was performed to decipher the differential gene expression and splicing modifications related to HTLV-1. Here we compared ATL-2 with MOLT-4, a non HTLV-1 derived leukemic T cell line and further compared with HBZ-induced modifications in an isogenic system composed by Jurkat T cells and stably HBZ transfected Jurkat derivatives. The endogenous HBZ interactome of ATL-2 cells identified 249 interactors covering three main clusters corresponding to protein families mainly involved in mRNA splicing, nonsense-mediated RNA decay (NMD) and JAK-STAT signaling pathway. Here we analyzed in detail the cluster involved in RNA splicing. RNAseq analysis showed that HBZ specifically altered the transcription of many genes, including crucial oncogenes, by affecting different splicing events. Consistently, the two RNA helicases, members of the RNA splicing family, DDX5 and its paralog DDX17, recently shown to be involved in alternative splicing of cellular genes after NF-κB activation by HTLV-1 Tax-1, interacted and partially co-localized with HBZ. For the first time, a complete picture of the endogenous HBZ interactome was elucidated. The wide interaction of HBZ with molecules involved in RNA splicing and the subsequent transcriptome alteration strongly suggests an unprecedented complex role of the viral oncogene in the establishment of the leukemic state.
Keywords: ATL; HBZ; HTLV-1; alternative splicing; interactome; protein network.
Copyright © 2022 Shallak, Alberio, Fasano, Monti, Iacobucci, Ladet, Mortreux, Accolla and Forlani.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Novel Interactions between the Human T-Cell Leukemia Virus Type 1 Antisense Protein HBZ and the SWI/SNF Chromatin Remodeling Family: Implications for Viral Life Cycle.J Virol. 2019 Jul 30;93(16):e00412-19. doi: 10.1128/JVI.00412-19. Print 2019 Aug 15. J Virol. 2019. PMID: 31142665 Free PMC article.
-
Localization, quantification and interaction with host factors of endogenous HTLV-1 HBZ protein in infected cells and ATL.Retrovirology. 2015 Jul 4;12:59. doi: 10.1186/s12977-015-0186-0. Retrovirology. 2015. PMID: 26140924 Free PMC article.
-
Characteristic expression of HTLV-1 basic zipper factor (HBZ) transcripts in HTLV-1 provirus-positive cells.Retrovirology. 2008 Apr 22;5:34. doi: 10.1186/1742-4690-5-34. Retrovirology. 2008. PMID: 18426605 Free PMC article.
-
HTLV-1 Infection and Adult T-Cell Leukemia/Lymphoma-A Tale of Two Proteins: Tax and HBZ.Viruses. 2016 Jun 16;8(6):161. doi: 10.3390/v8060161. Viruses. 2016. PMID: 27322308 Free PMC article. Review.
-
The Road to HTLV-1-Induced Leukemia by Following the Subcellular Localization of HTLV-1-Encoded HBZ Protein.Front Immunol. 2022 Jun 23;13:940131. doi: 10.3389/fimmu.2022.940131. eCollection 2022. Front Immunol. 2022. PMID: 35812456 Free PMC article. Review.
Cited by
-
Vital for Viruses: Intrinsically Disordered Proteins.J Mol Biol. 2023 Jun 1;435(11):167860. doi: 10.1016/j.jmb.2022.167860. Epub 2023 Jun 16. J Mol Biol. 2023. PMID: 37330280 Free PMC article. Review.
-
Unraveling the role of ZNF506 as a human PBS-pro-targeting protein for ERVP repression.Nucleic Acids Res. 2023 Oct 27;51(19):10309-10325. doi: 10.1093/nar/gkad731. Nucleic Acids Res. 2023. PMID: 37697430 Free PMC article.
-
Transcriptional co-activators: emerging roles in signaling pathways and potential therapeutic targets for diseases.Signal Transduct Target Ther. 2023 Nov 13;8(1):427. doi: 10.1038/s41392-023-01651-w. Signal Transduct Target Ther. 2023. PMID: 37953273 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

