Characterization of Macrophage-Tropic HIV-1 Infection of Central Nervous System Cells and the Influence of Inflammation
- PMID: 35975998
- PMCID: PMC9472603
- DOI: 10.1128/jvi.00957-22
Characterization of Macrophage-Tropic HIV-1 Infection of Central Nervous System Cells and the Influence of Inflammation
Abstract
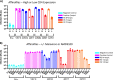
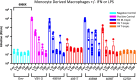
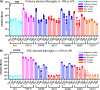
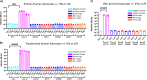
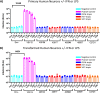
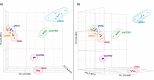
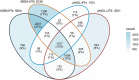
HIV-1 infection within the central nervous system (CNS) includes evolution of the virus, damaging inflammatory cascades, and the involvement of multiple cell types; however, our understanding of how Env tropism and inflammation can influence CNS infectivity is incomplete. In this study, we utilize macrophage-tropic and T cell-tropic HIV-1 Env proteins to establish accurate infection profiles for multiple CNS cells under basal and interferon alpha (IFN-α) or lipopolysaccharide (LPS)-induced inflammatory states. We found that macrophage-tropic viruses confer entry advantages in primary myeloid cells, including monocyte-derived macrophage, microglia, and induced pluripotent stem cell (iPSC)-derived microglia. However, neither macrophage-tropic or T cell-tropic HIV-1 Env proteins could mediate infection of astrocytes or neurons, and infection was not potentiated by induction of an inflammatory state in these cells. Additionally, we found that IFN-α and LPS restricted replication in myeloid cells, and IFN-α treatment prior to infection with vesicular stomatitis virus G protein (VSV G) Envs resulted in a conserved antiviral response across all CNS cell types. Further, using RNA sequencing (RNA-seq), we found that only myeloid cells express HIV-1 entry receptor/coreceptor transcripts at a significant level and that these transcripts in select cell types responded only modestly to inflammatory signals. We profiled the transcriptional response of multiple CNS cells to inflammation and found 57 IFN-induced genes that were differentially expressed across all cell types. Taken together, these data focus attention on the cells in the CNS that are truly permissive to HIV-1, further highlight the role of HIV-1 Env evolution in mediating infection in the CNS, and point to limitations in using model cell types versus primary cells to explore features of virus-host interaction. IMPORTANCE The major feature of HIV-1 pathogenesis is the induction of an immunodeficient state in the face of an enhanced state of inflammation. However, for many of those infected, there can be an impact on the central nervous system (CNS) resulting in a wide range of neurocognitive defects. Here, we use a highly sensitive and quantitative assay for viral infectivity to explore primary and model cell types of the brain for their susceptibility to infection using viral entry proteins derived from the CNS. In addition, we examine the ability of an inflammatory state to alter infectivity of these cells. We find that myeloid cells are the only cell types in the CNS that can be infected and that induction of an inflammatory state negatively impacts viral infection across all cell types.
Keywords: HIV-1; HIV-1 CNS inflammation; HIV-1 Env evolution; HIV-1 astrocyte infection; HIV-1 microglia infection; HIV-associated neurocognitive disorders; macrophage-tropism; neuroHIV.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Phenotypic Correlates of HIV-1 Macrophage Tropism.J Virol. 2015 Nov;89(22):11294-311. doi: 10.1128/JVI.00946-15. Epub 2015 Sep 2. J Virol. 2015. PMID: 26339058 Free PMC article.
-
Stoichiometry for entry and binding properties of the Env protein of R5 T cell-tropic HIV-1 and its evolutionary variant of macrophage-tropic HIV-1.mBio. 2024 Apr 10;15(4):e0032124. doi: 10.1128/mbio.00321-24. Epub 2024 Mar 1. mBio. 2024. PMID: 38426750 Free PMC article.
-
HIV-1 R5 Macrophage-Tropic Envelope Glycoprotein Trimers Bind CD4 with High Affinity, while the CD4 Binding Site on Non-macrophage-tropic, T-Tropic R5 Envelopes Is Occluded.J Virol. 2018 Jan 2;92(2):e00841-17. doi: 10.1128/JVI.00841-17. Print 2018 Jan 15. J Virol. 2018. PMID: 29118121 Free PMC article.
-
HIV-1 target cells in the CNS.J Neurovirol. 2015 Jun;21(3):276-89. doi: 10.1007/s13365-014-0287-x. Epub 2014 Sep 19. J Neurovirol. 2015. PMID: 25236812 Free PMC article. Review.
-
Viral determinants of HIV-1 macrophage tropism.Viruses. 2011 Nov;3(11):2255-79. doi: 10.3390/v3112255. Epub 2011 Nov 15. Viruses. 2011. PMID: 22163344 Free PMC article. Review.
Cited by
-
Changes in cerebrospinal fluid proteins across the spectrum of untreated and treated chronic HIV-1 infection.PLoS Pathog. 2024 Sep 24;20(9):e1012470. doi: 10.1371/journal.ppat.1012470. eCollection 2024 Sep. PLoS Pathog. 2024. PMID: 39316609 Free PMC article.
-
Astrocytes: Role in pathogenesis and effect of commonly misused drugs in the HIV infected brain.Curr Res Neurobiol. 2023 Aug 29;5:100108. doi: 10.1016/j.crneur.2023.100108. eCollection 2023. Curr Res Neurobiol. 2023. PMID: 38020814 Free PMC article. Review.
-
Characterization of HIV variants from paired Cerebrospinal fluid and Plasma samples in primary microglia and CD4+ T-cells.J Neurovirol. 2024 Aug;30(4):380-392. doi: 10.1007/s13365-024-01207-w. Epub 2024 May 7. J Neurovirol. 2024. PMID: 38713307 Free PMC article.
-
Persistence of intact HIV-1 proviruses in the brain during antiretroviral therapy.Elife. 2023 Nov 8;12:RP89837. doi: 10.7554/eLife.89837. Elife. 2023. PMID: 37938115 Free PMC article.
-
Dysregulated neuroimmune interactions and sustained type I interferon signaling after human immunodeficiency virus type 1 infection of human iPSC derived microglia and cerebral organoids.bioRxiv [Preprint]. 2023 Oct 30:2023.10.25.563950. doi: 10.1101/2023.10.25.563950. bioRxiv. 2023. Update in: iScience. 2024 Mar 28;27(5):109628. doi: 10.1016/j.isci.2024.109628 PMID: 37961371 Free PMC article. Updated. Preprint.
References
-
- Eggers C, Arendt G, Hahn K, Husstedt IW, Maschke M, Neuen-Jacob E, Obermann M, Rosenkranz T, Schielke E, Straube E, German Association of Neuro-AIDS und Neuro-Infectiology (DGNANI). 2017. HIV-1-associated neurocognitive disorder: epidemiology, pathogenesis, diagnosis, and treatment. J Neurol 264:1715–1727. 10.1007/s00415-017-8503-2. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

