Loss of Nexmif results in the expression of phenotypic variability and loss of genomic integrity
- PMID: 35970867
- PMCID: PMC9378738
- DOI: 10.1038/s41598-022-17845-1
Loss of Nexmif results in the expression of phenotypic variability and loss of genomic integrity
Abstract
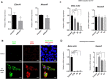
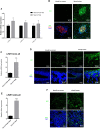
We identified two NEXMIF variants in two unrelated individuals with non-autoimmune diabetes and autistic traits, and investigated the expression of Nexmif in mouse and human pancreas and its function in pancreatic beta cells in vitro and in vivo. In insulin-secreting INS-1E cells, Nexmif expression increased strongly in response to oxidative stress. CRISPR Cas9-generated Nexmif knockout mice exhibited a reduced number of proliferating beta cells in pancreatic islets. RNA sequencing of pancreatic islets showed that the downregulated genes in Nexmif mutant islets are involved in stress response and the deposition of epigenetic marks. They include H3f3b, encoding histone H3.3, which is associated with the regulation of beta-cell proliferation and maintains genomic integrity by silencing transposable elements, particularly LINE1 elements. LINE1 activity has been associated with autism and neurodevelopmental disorders in which patients share characteristics with NEXMIF patients, and can cause genomic instability and genetic variation through retrotransposition. Nexmif knockout mice exhibited various other phenotypes. Mortality and phenotypic abnormalities increased in each generation in both Nexmif mutant and non-mutant littermates. In Nexmif mutant mice, LINE1 element expression was upregulated in the pancreas, brain, and testis, possibly inducing genomic instability in Nexmif mutant mice and causing phenotypic variability in their progeny.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
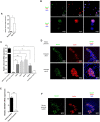

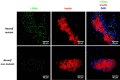
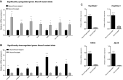
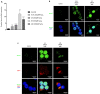
Similar articles
-
Heterozygous Nexmif female mice demonstrate mosaic NEXMIF expression, autism-like behaviors, and abnormalities in dendritic arborization and synaptogenesis.Heliyon. 2024 Jan 24;10(3):e24703. doi: 10.1016/j.heliyon.2024.e24703. eCollection 2024 Feb 15. Heliyon. 2024. PMID: 38322873 Free PMC article.
-
NEXMIF/KIDLIA Knock-out Mouse Demonstrates Autism-Like Behaviors, Memory Deficits, and Impairments in Synapse Formation and Function.J Neurosci. 2020 Jan 2;40(1):237-254. doi: 10.1523/JNEUROSCI.0222-19.2019. Epub 2019 Nov 8. J Neurosci. 2020. PMID: 31704787 Free PMC article.
-
P2Y1 purinergic receptor identified as a diabetes target in a small-molecule screen to reverse circadian β-cell failure.Elife. 2022 Feb 21;11:e75132. doi: 10.7554/eLife.75132. Elife. 2022. PMID: 35188462 Free PMC article.
-
Chronic exposure to leucine in vitro induces β-cell dysfunction in INS-1E cells and mouse islets.J Endocrinol. 2012 Oct;215(1):79-88. doi: 10.1530/JOE-12-0148. Epub 2012 Jul 13. J Endocrinol. 2012. PMID: 22798014
-
Clinical spectrum of KIAA2022/NEXMIF pathogenic variants in males and females: Report of three patients from Indian kindred with a review of published patients.Brain Dev. 2020 Oct;42(9):646-654. doi: 10.1016/j.braindev.2020.06.005. Epub 2020 Jun 27. Brain Dev. 2020. PMID: 32600841 Review.
Cited by
-
Heterozygous Nexmif female mice demonstrate mosaic NEXMIF expression, autism-like behaviors, and abnormalities in dendritic arborization and synaptogenesis.Heliyon. 2024 Jan 24;10(3):e24703. doi: 10.1016/j.heliyon.2024.e24703. eCollection 2024 Feb 15. Heliyon. 2024. PMID: 38322873 Free PMC article.
References
-
- Ishikawa T, Miyata S, Koyama Y, Yoshikawa K, Hattori T, Kumamoto N, et al. Transient expression of Xpn, an XLMR protein related to neurite extension, during brain development and participation in neurite outgrowth. Neuroscience. 2012;214:181–191. doi: 10.1016/j.neuroscience.2012.04.030. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

