A photo-switchable assay system for dendrite degeneration and repair in Drosophila melanogaster
- PMID: 35969739
- PMCID: PMC9407391
- DOI: 10.1073/pnas.2204577119
A photo-switchable assay system for dendrite degeneration and repair in Drosophila melanogaster
Abstract
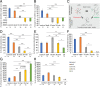
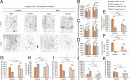
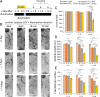
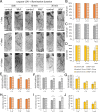
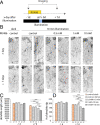
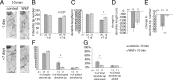
Neurodegeneration arising from aging, injury, or diseases has devastating health consequences. Whereas neuronal survival and axon degeneration have been studied extensively, much less is known about how neurodegeneration affects dendrites, in part due to the limited assay systems available. To develop an assay for dendrite degeneration and repair, we used photo-switchable caspase-3 (caspase-Light-Oxygen-Voltage-sensing [caspase-LOV]) in peripheral class 4 dendrite arborization (c4da) neurons to induce graded neurodegeneration by adjusting illumination duration during development and adulthood in Drosophila melanogaster. We found that both developing and mature c4da neurons were able to survive while sustaining mild neurodegeneration induced by moderate caspase-LOV activation. Further, we observed active dendrite addition and dendrite regeneration in developing and mature c4da neurons, respectively. Using this assay, we found that the mouse Wallerian degeneration slow (WldS) protein can protect c4da neurons from caspase-LOV-induced dendrite degeneration and cell death. Furthermore, our data show that WldS can reduce dendrite elimination without affecting dendrite addition. In summary, we successfully established a photo-switchable assay system in both developing and mature neurons and used WldS as a test case to study the mechanisms underlying dendrite regeneration and repair.
Keywords: Drosophila; Wallerian degeneration slow protein; dendrite degeneration; dendrite regeneration; dendrite repair.
Conflict of interest statement
The authors declare no competing interest.
Figures


Similar articles
-
Phagocytosis and self-destruction break down dendrites of Drosophila sensory neurons at distinct steps of Wallerian degeneration.Proc Natl Acad Sci U S A. 2022 Jan 25;119(4):e2111818119. doi: 10.1073/pnas.2111818119. Proc Natl Acad Sci U S A. 2022. PMID: 35058357 Free PMC article.
-
Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning.Neuron. 2006 Aug 3;51(3):283-90. doi: 10.1016/j.neuron.2006.07.014. Neuron. 2006. PMID: 16880123
-
Drosophila miR-87 promotes dendrite regeneration by targeting the transcriptional repressor Tramtrack69.PLoS Genet. 2020 Aug 7;16(8):e1008942. doi: 10.1371/journal.pgen.1008942. eCollection 2020 Aug. PLoS Genet. 2020. PMID: 32764744 Free PMC article.
-
Recent progress in dendritic pruning of Drosophila C4da sensory neurons.Open Biol. 2024 Jul;14(7):240059. doi: 10.1098/rsob.240059. Epub 2024 Jul 24. Open Biol. 2024. PMID: 39046196 Free PMC article. Review.
-
Out with the Old, In with the New: Dendrite Degeneration and Regeneration.In: Tran TS, Yaron A, editors. Wiring the Nervous System: Mechanisms of Axonal and Dendritic Remodelling in Health and Disease. 1st edition. Abingdon: River Publishers; 2024 Jan 31. Chapter 3. In: Tran TS, Yaron A, editors. Wiring the Nervous System: Mechanisms of Axonal and Dendritic Remodelling in Health and Disease. 1st edition. Abingdon: River Publishers; 2024 Jan 31. Chapter 3. PMID: 38536946 Free Books & Documents. Review.
Cited by
-
Motor neuron-specific RhoA knockout delays degeneration and promotes regeneration of dendrites in spinal ventral horn after brachial plexus injury.Neural Regen Res. 2023 Dec;18(12):2757-2761. doi: 10.4103/1673-5374.373657. Neural Regen Res. 2023. PMID: 37449641 Free PMC article.
References
-
- Kulkarni V. A., Firestein B. L., The dendritic tree and brain disorders. Mol. Cell. Neurosci. 50, 10–20 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

