Influence of single-nucleotide polymorphisms in TLR3 (rs3775291) and TLR9 (rs352139) on the risk of CMV infection in kidney transplant recipients
- PMID: 35967300
- PMCID: PMC9374175
- DOI: 10.3389/fimmu.2022.929995
Influence of single-nucleotide polymorphisms in TLR3 (rs3775291) and TLR9 (rs352139) on the risk of CMV infection in kidney transplant recipients
Abstract
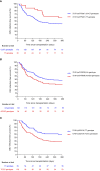
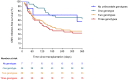
Risk stratification for cytomegalovirus (CMV) infection after kidney transplantation (KT) remains to be determined. Since endosomal toll-like receptors (TLRs) are involved in viral sensing, we investigated the impact of common single-nucleotide polymorphisms (SNPs) located within TLR3 and TLR9 genes on the occurrence of overall and high-level (≥1,000 IU/ml) CMV infection in a cohort of 197 KT recipients. Homozygous carriers of the minor allele of TLR3 (rs3775291) had higher infection-free survival compared with reference allele carriers (60.0% for TT versus 42.3% for CC/CT genotypes; P-value = 0.050). Decreased infection-free survival was observed with the minor allele of TLR9 (rs352139) (38.2% for TC/CC versus 59.3% for TT genotypes; P-value = 0.004). After multivariable adjustment, the recessive protective effect of the TLR3 (rs3775291) TT genotype was confirmed (adjusted hazard ratio [aHR]: 0.327; 95% CI: 0.167-0.642; P-value = 0.001), as was the dominant risk-conferring effect of TLR9 (rs352139) TC/CC genotypes (aHR: 1.865; 95% CI: 1.170-2.972; P-value = 0.009). Carriers of the TLR9 (rs352139) TC/CC genotypes showed lower CMV-specific interferon-γ-producing CD4+ T-cell counts measured by intracellular cytokine staining compared with the TT genotype (median of 0.2 versus 0.7 cells/μl; P-value = 0.003). In conclusion, TLR3/TLR9 genotyping may inform CMV infection risk after KT.
Keywords: cytomegalovirus; kidney transplantation; single-nucleotide polymorphism (SNP); toll-like receptor 3 (TLR3); toll-like receptor 9 (TLR9).
Copyright © 2022 Redondo, Rodríguez-Goncer, Parra, Ruiz-Merlo, López-Medrano, González, Polanco, Trujillo, Hernández, San Juan, Andrés, Aguado and Fernández-Ruiz.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
The Relationship between TLR3 rs3775291 Polymorphism and Infectious Diseases: A Meta-Analysis of Case-Control Studies.Genes (Basel). 2023 Jun 21;14(7):1311. doi: 10.3390/genes14071311. Genes (Basel). 2023. PMID: 37510216 Free PMC article. Review.
-
Association between individual and combined SNPs in genes related to innate immunity and incidence of CMV infection in seropositive kidney transplant recipients.Am J Transplant. 2015 May;15(5):1323-35. doi: 10.1111/ajt.13107. Epub 2015 Mar 16. Am J Transplant. 2015. PMID: 25777542
-
The TLR9 2848C/T Polymorphism Is Associated with the CMV DNAemia among HIV/CMV Co-Infected Patients.Cells. 2021 Sep 8;10(9):2360. doi: 10.3390/cells10092360. Cells. 2021. PMID: 34572011 Free PMC article.
-
Toll-like receptors genes polymorphisms and the occurrence of HCMV infection among pregnant women.Virol J. 2017 Mar 24;14(1):64. doi: 10.1186/s12985-017-0730-8. Virol J. 2017. PMID: 28340580 Free PMC article.
-
Host genetics and susceptibility to congenital and childhood cytomegalovirus infection: a systematic review.Croat Med J. 2016 Aug 31;57(4):321-30. doi: 10.3325/cmj.2016.57.321. Croat Med J. 2016. PMID: 27586547 Free PMC article. Review.
Cited by
-
Prediction of herpes virus infections after solid organ transplantation: a prospective study of immune function.Front Immunol. 2023 Jul 3;14:1183703. doi: 10.3389/fimmu.2023.1183703. eCollection 2023. Front Immunol. 2023. PMID: 37465673 Free PMC article.
-
The Relationship between TLR3 rs3775291 Polymorphism and Infectious Diseases: A Meta-Analysis of Case-Control Studies.Genes (Basel). 2023 Jun 21;14(7):1311. doi: 10.3390/genes14071311. Genes (Basel). 2023. PMID: 37510216 Free PMC article. Review.
-
Cytomegalovirus Cell-Mediated Immunity: Ready for Routine Use?Transpl Int. 2023 Nov 7;36:11963. doi: 10.3389/ti.2023.11963. eCollection 2023. Transpl Int. 2023. PMID: 38020746 Free PMC article. Review.
-
Impact of polymorphisms in genes orchestrating innate immune responses on replication kinetics of Torque teno virus after kidney transplantation.Front Genet. 2022 Nov 22;13:1069890. doi: 10.3389/fgene.2022.1069890. eCollection 2022. Front Genet. 2022. PMID: 36482892 Free PMC article.
-
The battle between host antiviral innate immunity and immune evasion by cytomegalovirus.Cell Mol Life Sci. 2024 Aug 9;81(1):341. doi: 10.1007/s00018-024-05369-y. Cell Mol Life Sci. 2024. PMID: 39120730 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

