Identification of Novel Inhibitors Targeting SGK1 via Ensemble-Based Virtual Screening Method, Biological Evaluation and Molecular Dynamics Simulation
- PMID: 35955763
- PMCID: PMC9369041
- DOI: 10.3390/ijms23158635
Identification of Novel Inhibitors Targeting SGK1 via Ensemble-Based Virtual Screening Method, Biological Evaluation and Molecular Dynamics Simulation
Abstract
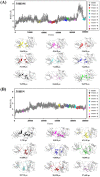
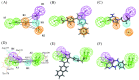
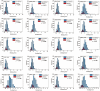
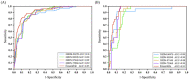
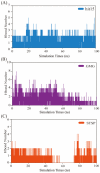
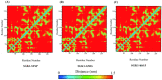
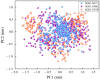
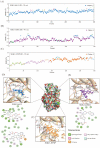
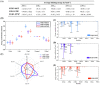
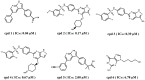
Serum and glucocorticoid-regulated kinase 1 (SGK1), as a serine threonine protein kinase of the AGC family, regulates different enzymes, transcription factors, ion channels, transporters, and cell proliferation and apoptosis. Inhibition of SGK1 is considered as a valuable approach for the treatment of various metabolic diseases. In this investigation, virtual screening methods, including pharmacophore models, Bayesian classifiers, and molecular docking, were combined to discover novel inhibitors of SGK1 from the database with 29,158 compounds. Then, the screened compounds were subjected to ADME/T, PAINS and drug-likeness analysis. Finally, 28 compounds with potential inhibition activity against SGK1 were selected for biological evaluation. The kinase inhibition activity test revealed that among these 28 hits, hit15 exhibited the highest inhibition activity against SGK1, which gave 44.79% inhibition rate at the concentration of 10 µM. In order to further investigate the interaction mechanism of hit15 and SGK1 at simulated physiological conditions, a molecular dynamics simulation was performed. The molecular dynamics simulation demonstrated that hit15 could bind to the active site of SGK1 and form stable interactions with key residues, such as Tyr178, ILE179, and VAL112. The binding free energy of the SGK1-hit15 was -48.90 kJ mol-1. Therefore, the identified hit15 with novel scaffold may be a promising lead compound for development of new SGK1 inhibitors for various diseases treatment.
Keywords: SGK1 inhibitor; biological evaluation; molecular dynamics; virtual screening.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Virtual Screening Approach to Identify High-Affinity Inhibitors of Serum and Glucocorticoid-Regulated Kinase 1 among Bioactive Natural Products: Combined Molecular Docking and Simulation Studies.Molecules. 2020 Feb 13;25(4):823. doi: 10.3390/molecules25040823. Molecules. 2020. PMID: 32070031 Free PMC article.
-
Discovery of novel S6K1 inhibitors by an ensemble-based virtual screening method and molecular dynamics simulation.J Mol Model. 2023 Mar 18;29(4):102. doi: 10.1007/s00894-023-05504-9. J Mol Model. 2023. PMID: 36933164
-
Discovery of microtubule stabilizers with novel scaffold structures based on virtual screening, biological evaluation, and molecular dynamics simulation.Chem Biol Interact. 2022 Jan 25;352:109784. doi: 10.1016/j.cbi.2021.109784. Epub 2021 Dec 18. Chem Biol Interact. 2022. PMID: 34932952
-
Therapeutic potential of serum and glucocorticoid inducible kinase inhibition.Expert Opin Investig Drugs. 2013 Jun;22(6):701-14. doi: 10.1517/13543784.2013.778971. Epub 2013 Mar 19. Expert Opin Investig Drugs. 2013. PMID: 23506284 Review.
-
Serum and glucocorticoid-regulated kinase 1: Structure, biological functions, and its inhibitors.Front Pharmacol. 2022 Nov 15;13:1036844. doi: 10.3389/fphar.2022.1036844. eCollection 2022. Front Pharmacol. 2022. PMID: 36457711 Free PMC article. Review.
Cited by
-
In silico exploration of deep-sea fungal metabolites as inhibitor of Ebola and Marburg VP35 and VP40.PLoS One. 2024 Jul 25;19(7):e0307579. doi: 10.1371/journal.pone.0307579. eCollection 2024. PLoS One. 2024. PMID: 39052567 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

