Impact of 100 LRRK2 variants linked to Parkinson's disease on kinase activity and microtubule binding
- PMID: 35950872
- PMCID: PMC9472821
- DOI: 10.1042/BCJ20220161
Impact of 100 LRRK2 variants linked to Parkinson's disease on kinase activity and microtubule binding
Abstract
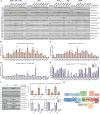
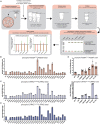
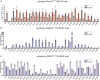
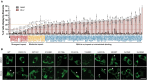
Mutations enhancing the kinase activity of leucine-rich repeat kinase-2 (LRRK2) cause Parkinson's disease (PD) and therapies that reduce LRRK2 kinase activity are being tested in clinical trials. Numerous rare variants of unknown clinical significance have been reported, but how the vast majority impact on LRRK2 function is unknown. Here, we investigate 100 LRRK2 variants linked to PD, including previously described pathogenic mutations. We identify 23 LRRK2 variants that robustly stimulate kinase activity, including variants within the N-terminal non-catalytic regions (ARM (E334K, A419V), ANK (R767H), LRR (R1067Q, R1325Q)), as well as variants predicted to destabilize the ROC:CORB interface (ROC (A1442P, V1447M), CORA (R1628P) CORB (S1761R, L1795F)) and COR:COR dimer interface (CORB (R1728H/L)). Most activating variants decrease LRRK2 biomarker site phosphorylation (pSer935/pSer955/pSer973), consistent with the notion that the active kinase conformation blocks their phosphorylation. We conclude that the impact of variants on kinase activity is best evaluated by deploying a cellular assay of LRRK2-dependent Rab10 substrate phosphorylation, compared with a biochemical kinase assay, as only a minority of activating variants (CORB (Y1699C, R1728H/L, S1761R) and kinase (G2019S, I2020T, T2031S)), enhance in vitro kinase activity of immunoprecipitated LRRK2. Twelve variants including several that activate LRRK2 and have been linked to PD, suppress microtubule association in the presence of a Type I kinase inhibitor (ARM (M712V), LRR (R1320S), ROC (A1442P, K1468E, S1508R), CORA (A1589S), CORB (Y1699C, R1728H/L) and WD40 (R2143M, S2350I, G2385R)). Our findings will stimulate work to better understand the mechanisms by which variants impact biology and provide rationale for variant carrier inclusion or exclusion in ongoing and future LRRK2 inhibitor clinical trials.
Keywords: G-proteins; Parkinson's disease; leucine-rich repeat kinase; signaling.
© 2022 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures





Similar articles
-
Clinical characteristics and pathophysiological properties of newly discovered LRRK2 variants associated with Parkinson's disease.Neurobiol Dis. 2024 Sep;199:106571. doi: 10.1016/j.nbd.2024.106571. Epub 2024 Jun 18. Neurobiol Dis. 2024. PMID: 38901781
-
LRRK2 phosphorylates moesin at threonine-558: characterization of how Parkinson's disease mutants affect kinase activity.Biochem J. 2007 Jul 15;405(2):307-17. doi: 10.1042/BJ20070209. Biochem J. 2007. PMID: 17447891 Free PMC article.
-
14-3-3 binding to LRRK2 is disrupted by multiple Parkinson's disease-associated mutations and regulates cytoplasmic localization.Biochem J. 2010 Sep 15;430(3):393-404. doi: 10.1042/BJ20100483. Biochem J. 2010. PMID: 20642453 Free PMC article.
-
Prevalence of ten LRRK2 variants in Parkinson's disease: A comprehensive review.Parkinsonism Relat Disord. 2022 May;98:103-113. doi: 10.1016/j.parkreldis.2022.05.012. Epub 2022 May 25. Parkinsonism Relat Disord. 2022. PMID: 35654702 Review.
-
Leucine-Rich Repeat Kinase (LRRK2) Genetics and Parkinson's Disease.Adv Neurobiol. 2017;14:3-30. doi: 10.1007/978-3-319-49969-7_1. Adv Neurobiol. 2017. PMID: 28353276 Review.
Cited by
-
Inhibition of Parkinson's disease-related LRRK2 by type I and type II kinase inhibitors: Activity and structures.Sci Adv. 2023 Dec;9(48):eadk6191. doi: 10.1126/sciadv.adk6191. Epub 2023 Dec 1. Sci Adv. 2023. PMID: 38039358 Free PMC article.
-
RAB12-LRRK2 complex suppresses primary ciliogenesis and regulates centrosome homeostasis in astrocytes.Nat Commun. 2024 Sep 29;15(1):8434. doi: 10.1038/s41467-024-52723-6. Nat Commun. 2024. PMID: 39343966 Free PMC article.
-
Comprehensive genetic screening of early-onset dementia patients in an Austrian cohort-suggesting new disease-contributing genes.Hum Genomics. 2023 Jun 17;17(1):55. doi: 10.1186/s40246-023-00499-z. Hum Genomics. 2023. PMID: 37330543 Free PMC article.
-
Elevated urine BMP phospholipids in LRRK2 and VPS35 mutation carriers with and without Parkinson's disease.NPJ Parkinsons Dis. 2023 Apr 4;9(1):52. doi: 10.1038/s41531-023-00482-4. NPJ Parkinsons Dis. 2023. PMID: 37015928 Free PMC article.
-
LRRK2 suppresses lysosome degradative activity in macrophages and microglia through MiT-TFE transcription factor inhibition.Proc Natl Acad Sci U S A. 2023 Aug;120(31):e2303789120. doi: 10.1073/pnas.2303789120. Epub 2023 Jul 24. Proc Natl Acad Sci U S A. 2023. PMID: 37487100 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

