Newly synthesized mRNA escapes translational repression during the acute phase of the mammalian unfolded protein response
- PMID: 35947624
- PMCID: PMC9365188
- DOI: 10.1371/journal.pone.0271695
Newly synthesized mRNA escapes translational repression during the acute phase of the mammalian unfolded protein response
Abstract
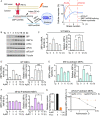
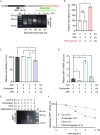
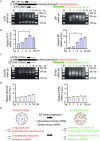
Endoplasmic Reticulum (ER) stress, caused by the accumulation of misfolded proteins in the ER, elicits a homeostatic mechanism known as the Unfolded Protein Response (UPR). The UPR reprograms gene expression to promote adaptation to chronic ER stress. The UPR comprises an acute phase involving inhibition of bulk protein synthesis and a chronic phase of transcriptional induction coupled with the partial recovery of protein synthesis. However, the role of transcriptional regulation in the acute phase of the UPR is not well understood. Here we analyzed the fate of newly synthesized mRNA encoding the protective and homeostatic transcription factor X-box binding protein 1 (XBP1) during this acute phase. We have previously shown that global translational repression induced by the acute UPR was characterized by decreased translation and increased stability of XBP1 mRNA. We demonstrate here that this stabilization is independent of new transcription. In contrast, we show XBP1 mRNA newly synthesized during the acute phase accumulates with long poly(A) tails and escapes translational repression. Inhibition of newly synthesized RNA polyadenylation during the acute phase decreased cell survival with no effect in unstressed cells. Furthermore, during the chronic phase of the UPR, levels of XBP1 mRNA with long poly(A) tails decreased in a manner consistent with co-translational deadenylation. Finally, additional pro-survival, transcriptionally-induced mRNAs show similar regulation, supporting the broad significance of the pre-steady state UPR in translational control during ER stress. We conclude that the biphasic regulation of poly(A) tail length during the UPR represents a previously unrecognized pro-survival mechanism of mammalian gene regulation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
A novel feedback loop regulates the response to endoplasmic reticulum stress via the cooperation of cytoplasmic splicing and mRNA translation.Mol Cell Biol. 2012 Mar;32(5):992-1003. doi: 10.1128/MCB.06665-11. Epub 2012 Jan 3. Mol Cell Biol. 2012. PMID: 22215619 Free PMC article.
-
miR-34c-5p modulates X-box-binding protein 1 (XBP1) expression during the adaptive phase of the unfolded protein response.FASEB J. 2019 Oct;33(10):11541-11554. doi: 10.1096/fj.201900600RR. Epub 2019 Jul 17. FASEB J. 2019. PMID: 31314593 Free PMC article.
-
Transcription of the NKG2D ligand MICA is suppressed by the IRE1/XBP1 pathway of the unfolded protein response through the regulation of E2F1.FASEB J. 2019 Mar;33(3):3481-3495. doi: 10.1096/fj.201801350RR. Epub 2018 Nov 19. FASEB J. 2019. PMID: 30452881
-
The multiple roles of the unfolded protein response regulator IRE1α in cancer.Mol Carcinog. 2019 Sep;58(9):1623-1630. doi: 10.1002/mc.23031. Epub 2019 Apr 30. Mol Carcinog. 2019. PMID: 31041814 Free PMC article. Review.
-
Emerging roles for the ER stress sensor IRE1α in metabolic regulation and disease.J Biol Chem. 2019 Dec 6;294(49):18726-18741. doi: 10.1074/jbc.REV119.007036. Epub 2019 Oct 30. J Biol Chem. 2019. PMID: 31666338 Free PMC article. Review.
Cited by
-
Detection of biofilm formation and antibiotics resistance of Staphylococcus spp. isolated from humans' and birds' oral cavities.Open Vet J. 2024 Sep;14(9):2215-2223. doi: 10.5455/OVJ.2024.v14.i9.9. Epub 2024 Sep 30. Open Vet J. 2024. PMID: 39553752 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

