SMAD4 and TGFβ are architects of inverse genetic programs during fate determination of antiviral CTLs
- PMID: 35942952
- PMCID: PMC9402230
- DOI: 10.7554/eLife.76457
SMAD4 and TGFβ are architects of inverse genetic programs during fate determination of antiviral CTLs
Abstract
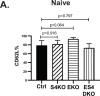
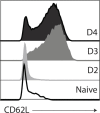
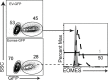
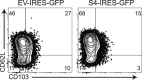
Transforming growth factor β (TGFβ) is an important differentiation factor for cytotoxic T lymphocytes (CTLs) and alters the expression levels of several of homing receptors during infection. SMAD4 is part of the canonical signaling network used by members of the transforming growth factor family. For this study, genetically modified mice were used to determine how SMAD4 and TGFβ receptor II (TGFβRII) participate in transcriptional programming of pathogen-specific CTLs. We show that these molecules are essential components of opposing signaling mechanisms, and cooperatively regulate a collection of genes that determine whether specialized populations of pathogen-specific CTLs circulate around the body, or settle in peripheral tissues. TGFβ uses a canonical SMAD-dependent signaling pathway to downregulate Eomesodermin (EOMES), KLRG1, and CD62L, while CD103 is induced. Conversely, in vivo and in vitro data show that EOMES, KLRG1, CX3CR1, and CD62L are positively regulated via SMAD4, while CD103 and Hobit are downregulated. Intravascular staining also shows that signaling via SMAD4 promotes formation of long-lived terminally differentiated CTLs that localize in the vasculature. Our data show that inflammatory molecules play a key role in lineage determination of pathogen-specific CTLs, and use SMAD-dependent signaling to alter the expression levels of multiple homing receptors and transcription factors with known functions during memory formation.
Keywords: CD8 T cell differentiation; CD8 T cells; CD8 memory; SMAD4; TGFβ; cytotoxic T lymphocyte; immunology; inflammation; influenza a virus; listeria monocytogenes; mouse; respiratory virus.
© 2022, Chandiran et al.
Conflict of interest statement
KC, JS, YH, EJ, ZU, JL, BM, SK, LC No competing interests declared
Figures





Similar articles
-
The diverse effects of transforming growth factor-β and SMAD signaling pathways during the CTL response.Front Immunol. 2023 Jun 23;14:1199671. doi: 10.3389/fimmu.2023.1199671. eCollection 2023. Front Immunol. 2023. PMID: 37426662 Free PMC article. Review.
-
Smad4 promotes differentiation of effector and circulating memory CD8 T cells but is dispensable for tissue-resident memory CD8 T cells.J Immunol. 2015 Mar 1;194(5):2407-14. doi: 10.4049/jimmunol.1402369. Epub 2015 Jan 30. J Immunol. 2015. PMID: 25637015 Free PMC article.
-
Smad4-Irf6 genetic interaction and TGFβ-mediated IRF6 signaling cascade are crucial for palatal fusion in mice.Development. 2013 Mar;140(6):1220-30. doi: 10.1242/dev.089615. Epub 2013 Feb 13. Development. 2013. PMID: 23406900 Free PMC article.
-
A TGFβ-Smad4-Fgf6 signaling cascade controls myogenic differentiation and myoblast fusion during tongue development.Development. 2012 May;139(9):1640-50. doi: 10.1242/dev.076653. Epub 2012 Mar 21. Development. 2012. PMID: 22438570 Free PMC article.
-
Bone morphogenetic proteins.Growth Factors. 2004 Dec;22(4):233-41. doi: 10.1080/08977190412331279890. Growth Factors. 2004. PMID: 15621726 Review.
Cited by
-
The diverse effects of transforming growth factor-β and SMAD signaling pathways during the CTL response.Front Immunol. 2023 Jun 23;14:1199671. doi: 10.3389/fimmu.2023.1199671. eCollection 2023. Front Immunol. 2023. PMID: 37426662 Free PMC article. Review.
-
TGF-β neutralization attenuates tumor residency of activated T cells to enhance systemic immunity in mice.iScience. 2024 Jul 15;27(8):110520. doi: 10.1016/j.isci.2024.110520. eCollection 2024 Aug 16. iScience. 2024. PMID: 39139402 Free PMC article.
-
CTLs Get SMAD When Pathogens Tell Them Where to Go.J Immunol. 2022 Sep 15;209(6):1025-1032. doi: 10.4049/jimmunol.2200345. J Immunol. 2022. PMID: 36130123 Free PMC article. Review.
-
Editorial: TGF-β and T cell biology.Front Immunol. 2023 Sep 5;14:1282656. doi: 10.3389/fimmu.2023.1282656. eCollection 2023. Front Immunol. 2023. PMID: 37736097 Free PMC article. No abstract available.
References
-
- Cao J, Zhang X, Wang Q, Qiu G, Hou C, Wang J, Cheng Q, Lan Y, Han H, Shen H, Zhang Y, Yang X, Shen B, Zhang J. Smad4 represses the generation of memory-precursor effector T cells but is required for the differentiation of central memory T cells. Cell Death & Disease. 2015;6:e1984. doi: 10.1038/cddis.2015.337. - DOI - PMC - PubMed
-
- Chang JT, Ciocca ML, Kinjyo I, Palanivel VR, McClurkin CE, Dejong CS, Mooney EC, Kim JS, Steinel NC, Oliaro J, Yin CC, Florea BI, Overkleeft HS, Berg LJ, Russell SM, Koretzky GA, Jordan MS, Reiner SL. Asymmetric proteasome segregation as a mechanism for unequal partitioning of the transcription factor T-bet during T lymphocyte division. Immunity. 2011;34:492–504. doi: 10.1016/j.immuni.2011.03.017. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

