Lipid Transport from Endoplasmic Reticulum to Autophagic Membranes
- PMID: 35940912
- PMCID: PMC9620852
- DOI: 10.1101/cshperspect.a041254
Lipid Transport from Endoplasmic Reticulum to Autophagic Membranes
Abstract
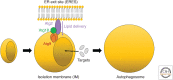
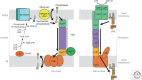
Autophagy is an intracellular degradation system involving de novo generation of autophagosomes, which function as a transporting vesicle of cytoplasmic components to lysosomes for degradation. Isolation membranes (IMs) or phagophores, the precursor membranes of autophagosomes, require millions of phospholipids to expand and transform into autophagosomes, with the endoplasmic reticulum (ER) being the primary lipid source. Recent advances in structural and biochemical studies of autophagy-related proteins have revealed their lipid transport activities: Atg2 at the interface between IM and ER possesses intermembrane lipid transfer activities, while Atg9 at IM and VMP1 and TMEM41B at ER possess lipid scrambling activities. In this review, we summarize recent advances in the establishment of the lipid transport activities of these proteins and their collaboration mechanisms for lipid transport between the ER and IM, and further discuss how unidirectional lipid transport from the ER to IM occurs during autophagosome formation.
Copyright © 2022 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures


Similar articles
-
Autophagosome formation in relation to the endoplasmic reticulum.J Biomed Sci. 2020 Oct 22;27(1):97. doi: 10.1186/s12929-020-00691-6. J Biomed Sci. 2020. PMID: 33087127 Free PMC article. Review.
-
--Atg9 interactions via its transmembrane domains are required for phagophore expansion during autophagy.Autophagy. 2023 May;19(5):1459-1478. doi: 10.1080/15548627.2022.2136340. Epub 2022 Nov 10. Autophagy. 2023. PMID: 36354155 Free PMC article.
-
A model for a partnership of lipid transfer proteins and scramblases in membrane expansion and organelle biogenesis.Proc Natl Acad Sci U S A. 2021 Apr 20;118(16):e2101562118. doi: 10.1073/pnas.2101562118. Proc Natl Acad Sci U S A. 2021. PMID: 33850023 Free PMC article.
-
Atg2: A novel phospholipid transfer protein that mediates de novo autophagosome biogenesis.Protein Sci. 2019 Jun;28(6):1005-1012. doi: 10.1002/pro.3623. Epub 2019 Apr 29. Protein Sci. 2019. PMID: 30993752 Free PMC article. Review.
-
Atg2 mediates direct lipid transfer between membranes for autophagosome formation.Nat Struct Mol Biol. 2019 Apr;26(4):281-288. doi: 10.1038/s41594-019-0203-4. Epub 2019 Mar 25. Nat Struct Mol Biol. 2019. PMID: 30911189
Cited by
-
Atl (atlastin) regulates mTor signaling and autophagy in Drosophila muscle through alteration of the lysosomal network.Autophagy. 2024 Jan;20(1):131-150. doi: 10.1080/15548627.2023.2249794. Epub 2023 Aug 30. Autophagy. 2024. PMID: 37649246 Free PMC article.
-
Mechanisms of autophagy-lysosome dysfunction in neurodegenerative diseases.Nat Rev Mol Cell Biol. 2024 Nov;25(11):926-946. doi: 10.1038/s41580-024-00757-5. Epub 2024 Aug 6. Nat Rev Mol Cell Biol. 2024. PMID: 39107446 Review.
References
-
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 182: 685–701. 10.1083/jcb.200803137 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
