Reversible structural changes in the influenza hemagglutinin precursor at membrane fusion pH
- PMID: 35939703
- PMCID: PMC9388137
- DOI: 10.1073/pnas.2208011119
Reversible structural changes in the influenza hemagglutinin precursor at membrane fusion pH
Abstract
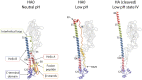
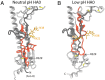
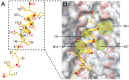
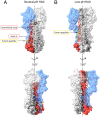
The subunits of the influenza hemagglutinin (HA) trimer are synthesized as single-chain precursors (HA0s) that are proteolytically cleaved into the disulfide-linked polypeptides HA1 and HA2. Cleavage is required for activation of membrane fusion at low pH, which occurs at the beginning of infection following transfer of cell-surface-bound viruses into endosomes. Activation results in extensive changes in the conformation of cleaved HA. To establish the overall contribution of cleavage to the mechanism of HA-mediated membrane fusion, we used cryogenic electron microscopy (cryo-EM) to directly image HA0 at neutral and low pH. We found extensive pH-induced structural changes, some of which were similar to those described for intermediates in the refolding of cleaved HA at low pH. They involve a partial extension of the long central coiled coil formed by melting of the preexisting secondary structure, threading it between the membrane-distal domains, and subsequent refolding as extended helices. The fusion peptide, covalently linked at its N terminus, adopts an amphipathic helical conformation over part of its length and is repositioned and packed against a complementary surface groove of conserved residues. Furthermore, and in contrast to cleaved HA, the changes in HA0 structure at low pH are reversible on reincubation at neutral pH. We discuss the implications of covalently restricted HA0 refolding for the cleaved HA conformational changes that mediate membrane fusion and for the action of antiviral drug candidates and cross-reactive anti-HA antibodies that can block influenza infectivity.
Keywords: cryo-EM; hemagglutinin; influenza; membrane fusion; protein folding.
Conflict of interest statement
The authors declare no competing interest.
Figures

Similar articles
-
Intermonomer Interactions in Hemagglutinin Subunits HA1 and HA2 Affecting Hemagglutinin Stability and Influenza Virus Infectivity.J Virol. 2015 Oct;89(20):10602-11. doi: 10.1128/JVI.00939-15. Epub 2015 Aug 12. J Virol. 2015. PMID: 26269180 Free PMC article.
-
Structural transitions in influenza haemagglutinin at membrane fusion pH.Nature. 2020 Jul;583(7814):150-153. doi: 10.1038/s41586-020-2333-6. Epub 2020 May 27. Nature. 2020. PMID: 32461688 Free PMC article.
-
Structural intermediates in the low pH-induced transition of influenza hemagglutinin.PLoS Pathog. 2020 Nov 30;16(11):e1009062. doi: 10.1371/journal.ppat.1009062. eCollection 2020 Nov. PLoS Pathog. 2020. PMID: 33253316 Free PMC article.
-
Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin.Annu Rev Biochem. 2000;69:531-69. doi: 10.1146/annurev.biochem.69.1.531. Annu Rev Biochem. 2000. PMID: 10966468 Review.
-
Composition and functions of the influenza fusion peptide.Protein Pept Lett. 2009;16(7):766-78. doi: 10.2174/092986609788681715. Protein Pept Lett. 2009. PMID: 19601906 Review.
Cited by
-
Rational design of lipid nanoparticles: overcoming physiological barriers for selective intracellular mRNA delivery.Curr Opin Chem Biol. 2024 Aug;81:102499. doi: 10.1016/j.cbpa.2024.102499. Epub 2024 Jul 13. Curr Opin Chem Biol. 2024. PMID: 38996568 Review.
-
Intradermal Immunization of Soluble Influenza HA Derived from a Lethal Virus Induces High Magnitude and Breadth of Antibody Responses and Provides Complete Protection In Vivo.Vaccines (Basel). 2023 Mar 31;11(4):780. doi: 10.3390/vaccines11040780. Vaccines (Basel). 2023. PMID: 37112692 Free PMC article.
-
Structural characterisation of hemagglutinin from seven Influenza A H1N1 strains reveal diversity in the C05 antibody recognition site.Sci Rep. 2023 Apr 28;13(1):6940. doi: 10.1038/s41598-023-33529-w. Sci Rep. 2023. PMID: 37117205 Free PMC article.
-
Disulfide-stabilized trimeric hemagglutinin ectodomains provide enhanced heterologous influenza protection.Emerg Microbes Infect. 2024 Dec;13(1):2389095. doi: 10.1080/22221751.2024.2389095. Epub 2024 Aug 18. Emerg Microbes Infect. 2024. PMID: 39101691 Free PMC article.
-
Engineering a cleaved, prefusion-stabilized influenza B virus hemagglutinin by identification and locking of all six pH switches.PNAS Nexus. 2024 Oct 11;3(10):pgae462. doi: 10.1093/pnasnexus/pgae462. eCollection 2024 Oct. PNAS Nexus. 2024. PMID: 39445049 Free PMC article.
References
-
- Klenk H. D., Rott R., Orlich M., Blödorn J., Activation of influenza A viruses by trypsin treatment. Virology 68, 426–439 (1975). - PubMed
-
- Laver W. G., Separation of two polypeptide chains from the hemagglutinin subunit of influenza virus. Virology 45, 275–288 (1971). - PubMed
-
- Lazarowitz S. G., Choppin P. W., Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology 68, 440–454 (1975). - PubMed
-
- Skehel J. J., Schild G. C., The polypeptide composition of influenza A viruses. Virology 44, 396–408 (1971). - PubMed
-
- Klenk H. D., Rott R., Orlich M., Further studies on the activation of influenza virus by proteolytic cleavage of the haemagglutinin. J. Gen. Virol. 36, 151–161 (1977). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

