PTX3 structure determination using a hybrid cryoelectron microscopy and AlphaFold approach offers insights into ligand binding and complement activation
- PMID: 35939690
- PMCID: PMC9388099
- DOI: 10.1073/pnas.2208144119
PTX3 structure determination using a hybrid cryoelectron microscopy and AlphaFold approach offers insights into ligand binding and complement activation
Abstract
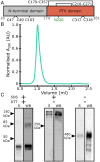
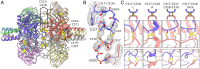
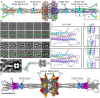
Pattern recognition molecules (PRMs) form an important part of innate immunity, where they facilitate the response to infections and damage by triggering processes such as inflammation. The pentraxin family of soluble PRMs comprises long and short pentraxins, with the former containing unique N-terminal regions unrelated to other proteins or each other. No complete high-resolution structural information exists about long pentraxins, unlike the short pentraxins, where there is an abundance of both X-ray and cryoelectron microscopy (cryo-EM)-derived structures. This study presents a high-resolution structure of the prototypical long pentraxin, PTX3. Cryo-EM yielded a 2.5-Å map of the C-terminal pentraxin domains that revealed a radically different quaternary structure compared to other pentraxins, comprising a glycosylated D4 symmetrical octameric complex stabilized by an extensive disulfide network. The cryo-EM map indicated α-helices that extended N terminal of the pentraxin domains that were not fully resolved. AlphaFold was used to predict the remaining N-terminal structure of the octameric PTX3 complex, revealing two long tetrameric coiled coils with two hinge regions, which was validated using classification of cryo-EM two-dimensional averages. The resulting hybrid cryo-EM/AlphaFold structure allowed mapping of ligand binding sites, such as C1q and fibroblast growth factor-2, as well as rationalization of previous biochemical data. Given the relevance of PTX3 in conditions ranging from COVID-19 prognosis, cancer progression, and female infertility, this structure could be used to inform the understanding and rational design of therapies for these disorders and processes.
Keywords: AlphaFold; COVID19; Long pentraxin; complement; cryoEM.
Conflict of interest statement
The authors declare no competing interest.
Figures


Similar articles
-
Structural insights into the biological functions of the long pentraxin PTX3.Front Immunol. 2023 Oct 9;14:1274634. doi: 10.3389/fimmu.2023.1274634. eCollection 2023. Front Immunol. 2023. PMID: 37885881 Free PMC article. Review.
-
Multimer formation and ligand recognition by the long pentraxin PTX3. Similarities and differences with the short pentraxins C-reactive protein and serum amyloid P component.J Biol Chem. 1997 Dec 26;272(52):32817-23. doi: 10.1074/jbc.272.52.32817. J Biol Chem. 1997. PMID: 9407058
-
The long pentraxin PTX3: a paradigm for humoral pattern recognition molecules.Ann N Y Acad Sci. 2013 May;1285:1-14. doi: 10.1111/nyas.12043. Epub 2013 Mar 25. Ann N Y Acad Sci. 2013. PMID: 23527487 Review.
-
Pentraxins in innate immunity: from C-reactive protein to the long pentraxin PTX3.J Clin Immunol. 2008 Jan;28(1):1-13. doi: 10.1007/s10875-007-9126-7. Epub 2007 Sep 9. J Clin Immunol. 2008. PMID: 17828584 Review.
-
PTX3 as a paradigm for the interaction of pentraxins with the complement system.Semin Immunol. 2013 Feb;25(1):79-85. doi: 10.1016/j.smim.2013.05.002. Epub 2013 Jun 6. Semin Immunol. 2013. PMID: 23747040 Review.
Cited by
-
Soluble Human Lectins at the Host-Microbe Interface.Annu Rev Biochem. 2024 Aug;93(1):565-601. doi: 10.1146/annurev-biochem-062917-012322. Epub 2024 Jul 2. Annu Rev Biochem. 2024. PMID: 38640018 Review.
-
Long Pentraxin 3 as a New Biomarker for Diagnosis of Hip and Knee Periprosthetic Joint Infections.J Clin Med. 2023 Jan 29;12(3):1055. doi: 10.3390/jcm12031055. J Clin Med. 2023. PMID: 36769703 Free PMC article.
-
Genetic Deficiency of the Long Pentraxin 3 Affects Osteogenesis and Osteoclastogenesis in Homeostatic and Inflammatory Conditions.Int J Mol Sci. 2023 Nov 23;24(23):16648. doi: 10.3390/ijms242316648. Int J Mol Sci. 2023. PMID: 38068970 Free PMC article.
-
Functional variants of the pentraxin 3 gene are associated with the metastasis and progression of prostate cancer.J Cell Mol Med. 2024 Aug;28(16):e70041. doi: 10.1111/jcmm.70041. J Cell Mol Med. 2024. PMID: 39187920 Free PMC article.
-
Structural insights into the biological functions of the long pentraxin PTX3.Front Immunol. 2023 Oct 9;14:1274634. doi: 10.3389/fimmu.2023.1274634. eCollection 2023. Front Immunol. 2023. PMID: 37885881 Free PMC article. Review.
References
-
- Garlanda C., Bottazzi B., Bastone A., Mantovani A., Pentraxins at the crossroads between innate immunity, inflammation, matrix deposition, and female fertility. Annu. Rev. Immunol. 23, 337–366 (2005). - PubMed
-
- Reading P. C., et al. , Antiviral activity of the long chain pentraxin PTX3 against influenza viruses. J. Immunol. 180, 3391–3398 (2008). - PubMed
-
- Salustri A., et al. , PTX3 plays a key role in the organization of the cumulus oophorus extracellular matrix and in in vivo fertilization. Development 131, 1577–1586 (2004). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

