Genome-Scale Metabolic Modeling Combined with Transcriptome Profiling Provides Mechanistic Understanding of Streptococcus thermophilus CH8 Metabolism
- PMID: 35924931
- PMCID: PMC9477255
- DOI: 10.1128/aem.00780-22
Genome-Scale Metabolic Modeling Combined with Transcriptome Profiling Provides Mechanistic Understanding of Streptococcus thermophilus CH8 Metabolism
Abstract
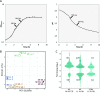
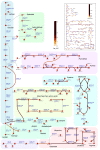
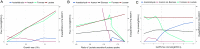
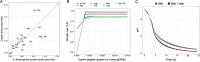
Streptococcus thermophilus is a lactic acid bacterium adapted toward growth in milk and is a vital component of starter cultures for milk fermentation. Here, we combine genome-scale metabolic modeling and transcriptome profiling to obtain novel metabolic insights into this bacterium. Notably, a refined genome-scale metabolic model (GEM) accurately representing S. thermophilus CH8 metabolism was developed. Modeling the utilization of casein as a nitrogen source revealed an imbalance in amino acid supply and demand, resulting in growth limitation due to the scarcity of specific amino acids, in particular sulfur amino acids. Growth experiments in milk corroborated this finding. A subtle interdependency of the redox balance and the secretion levels of the key metabolites lactate, formate, acetoin, and acetaldehyde was furthermore identified with the modeling approach, providing a mechanistic understanding of the factors governing the secretion product profile. As a potential effect of high expression of arginine biosynthesis genes, a moderate secretion of ornithine was observed experimentally, augmenting the proposed hypothesis of ornithine/putrescine exchange as part of the protocooperative interaction between S. thermophilus and Lactobacillus delbrueckii subsp. bulgaricus in yogurt. This study provides a foundation for future community modeling of food fermentations and rational development of starter strains with improved functionality. IMPORTANCE Streptococcus thermophilus is one the main organisms involved in the fermentation of milk and, increasingly, also in the fermentation of plant-based foods. The construction of a functional high-quality genome-scale metabolic model, in conjunction with in-depth transcriptome profiling with a focus on metabolism, provides a valuable resource for the improved understanding of S. thermophilus physiology. An example is the model-based prediction of the most significant route of synthesis for the characteristic yogurt flavor compound acetaldehyde and identification of metabolic principles governing the synthesis of other flavor compounds. Moreover, the systematic assessment of amino acid supply and demand during growth in milk provides insights into the key challenges related to nitrogen metabolism that is imposed on S. thermophilus and any other organism associated with the milk niche.
Keywords: RNA-Seq; Streptococcus thermophilus; food fermentation; genome-scale metabolic modeling; microbial biotechnology; microbial metabolism.
Conflict of interest statement
The authors declare a conflict of interest. M.H.R., P.G., M.L.J., A.G., A.R.N., and A.A.Z. are present or previous employees at Chr. Hansen A/S, a global supplier of food cultures and enzymes. The authors' views presented in this manuscript, however, are solely based on scientific grounds and do not reflect the commercial interests of their employer.
Figures

Similar articles
-
Comparative transcriptomic analysis of the flavor production mechanism in yogurt by traditional starter strains.J Dairy Sci. 2024 Aug;107(8):5402-5415. doi: 10.3168/jds.2023-24328. Epub 2024 Feb 7. J Dairy Sci. 2024. PMID: 38331185
-
Mixed-culture transcriptome analysis reveals the molecular basis of mixed-culture growth in Streptococcus thermophilus and Lactobacillus bulgaricus.Appl Environ Microbiol. 2010 Dec;76(23):7775-84. doi: 10.1128/AEM.01122-10. Epub 2010 Oct 1. Appl Environ Microbiol. 2010. PMID: 20889781 Free PMC article.
-
Effect of lactose hydrolysis on the milk-fermenting properties of Lactobacillus delbrueckii ssp. bulgaricus 2038 and Streptococcus thermophilus 1131.J Dairy Sci. 2021 Feb;104(2):1454-1464. doi: 10.3168/jds.2020-19244. Epub 2020 Dec 11. J Dairy Sci. 2021. PMID: 33309355
-
Harnessing the metabolic potential of Streptococcus thermophilus for new biotechnological applications.Curr Opin Biotechnol. 2020 Feb;61:142-152. doi: 10.1016/j.copbio.2019.12.019. Epub 2020 Jan 13. Curr Opin Biotechnol. 2020. PMID: 31945498 Review.
-
[Gene regulation to lactic acid bacteria for increasing production of flavor metabolite].Wei Sheng Wu Xue Bao. 2007 Dec;47(6):1105-9. Wei Sheng Wu Xue Bao. 2007. PMID: 18271275 Review. Chinese.
Cited by
-
Reconstruction and Analysis of a Genome-Scale Metabolic Model of Acinetobacter lwoffii.Int J Mol Sci. 2024 Aug 28;25(17):9321. doi: 10.3390/ijms25179321. Int J Mol Sci. 2024. PMID: 39273268 Free PMC article.
-
A Two-Compartment Fermentation System to Quantify Strain-Specific Interactions in Microbial Co-Cultures.Bioengineering (Basel). 2023 Jan 11;10(1):103. doi: 10.3390/bioengineering10010103. Bioengineering (Basel). 2023. PMID: 36671675 Free PMC article.
-
Microbial interactions shape cheese flavour formation.Nat Commun. 2023 Dec 21;14(1):8348. doi: 10.1038/s41467-023-41059-2. Nat Commun. 2023. PMID: 38129392 Free PMC article.
-
The Impact of Bioactive Molecules from Probiotics on Child Health: A Comprehensive Review.Nutrients. 2024 Oct 30;16(21):3706. doi: 10.3390/nu16213706. Nutrients. 2024. PMID: 39519539 Free PMC article. Review.
-
Differential Amino Acid Uptake and Depletion in Mono-Cultures and Co-Cultures of Streptococcus thermophilus and Lactobacillus delbrueckii subsp. bulgaricus in a Novel Semi-Synthetic Medium.Microorganisms. 2022 Sep 1;10(9):1771. doi: 10.3390/microorganisms10091771. Microorganisms. 2022. PMID: 36144373 Free PMC article.
References
-
- Hols P, Hancy F, Fontaine L, Grossiord B, Prozzi D, Leblond-Bourget N, Decaris B, Bolotin A, Delorme C, Dusko Ehrlich S, Guédon E, Monnet V, Renault P, Kleerebezem M. 2005. New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol Rev 29:435–463. 10.1016/j.femsre.2005.04.008. - DOI - PubMed
-
- Orla-Jensen S. 1919. The lactic acid bacteria. A. F. Høst og Søn, Copenhagen, Denmark.
-
- Bolotin A, Quinquis B, Renault P, Sorokin A, Ehrlich SD, Kulakauskas S, Lapidus A, Goltsman E, Mazur M, Pusch GD, Fonstein M, Overbeek R, Kyprides N, Purnelle B, Prozzi D, Ngui K, Masuy D, Hancy F, Burteau S, Boutry M, Delcour J, Goffeau A, Hols P. 2004. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat Biotechnol 22:1554–1558. 10.1038/nbt1034. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

